N-terminal acetylation of the yeast Derlin Der1 is essential for Hrd1 ubiquitin-ligase activity toward luminal ER substrates
- PMID: 23363603
- PMCID: PMC3608499
- DOI: 10.1091/mbc.E12-11-0838
N-terminal acetylation of the yeast Derlin Der1 is essential for Hrd1 ubiquitin-ligase activity toward luminal ER substrates
Abstract
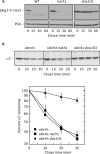
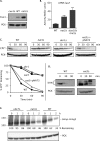
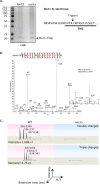
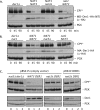
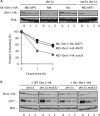
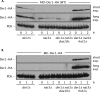
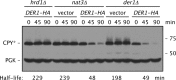
Two conserved ubiquitin ligases, Hrd1 and Doa10, mediate most endoplasmic reticulum-associated protein degradation (ERAD) in yeast. Degradation signals (degrons) recognized by these ubiquitin ligases remain poorly characterized. Doa10 recognizes the Deg1 degron from the MATα2 transcription factor. We previously found that deletion of the gene (NAT3) encoding the catalytic subunit of the NatB N-terminal acetyltransferase weakly stabilized a Deg1-fusion protein. By contrast, a recent analysis of several MATα2 derivatives suggested that N-terminal acetylation of these proteins by NatB was crucial for recognition by Doa10. We now analyze endogenous MATα2 degradation in cells lacking NatB and observe minimal perturbation relative to wild-type cells. However, NatB mutation strongly impairs degradation of ER-luminal Hrd1 substrates. This unexpected defect derives from a failure of Der1, a Hrd1 complex subunit, to be N-terminally acetylated in NatB mutant yeast. We retargeted Der1 to another acetyltransferase to show that it is the only ERAD factor requiring N-terminal acetylation. Preventing Der1 acetylation stimulates its proteolysis via the Hrd1 pathway, at least partially accounting for the ERAD defect observed in the absence of NatB. These results reveal an important role for N-terminal acetylation in controlling Hrd1 ligase activity toward a specific class of ERAD substrates.
Figures
References
-
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER- associated degradation. Nat Cell Biol. 2001;3:24–29. - PubMed
-
- Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. - PubMed
-
- Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. - PubMed
-
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell. 1993;74:357–369. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

