TORC1-regulated protein kinase Npr1 phosphorylates Orm to stimulate complex sphingolipid synthesis
- PMID: 23363605
- PMCID: PMC3596256
- DOI: 10.1091/mbc.E12-10-0753
TORC1-regulated protein kinase Npr1 phosphorylates Orm to stimulate complex sphingolipid synthesis
Abstract
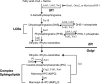
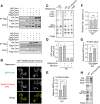
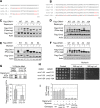
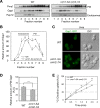
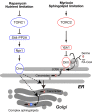
The evolutionarily conserved Orm1 and Orm2 proteins mediate sphingolipid homeostasis. However, the homologous Orm proteins and the signaling pathways modulating their phosphorylation and function are incompletely characterized. Here we demonstrate that inhibition of nutrient-sensitive target of rapamycin complex 1 (TORC1) stimulates Orm phosphorylation and synthesis of complex sphingolipids in Saccharomyces cerevisiae. TORC1 inhibition activates the kinase Npr1 that directly phosphorylates and activates the Orm proteins. Npr1-phosphorylated Orm1 and Orm2 stimulate de novo synthesis of complex sphingolipids downstream of serine palmitoyltransferase. Complex sphingolipids in turn stimulate plasma membrane localization and activity of the nutrient scavenging general amino acid permease 1. Thus activation of Orm and complex sphingolipid synthesis upon TORC1 inhibition is a physiological response to starvation.
Figures


References
-
- Arndt KT, Styles CA, Fink GR. A suppressor of a HIS4 transcriptional defect encodes a protein with homology to the catalytic subunit of protein phosphatases. Cell. 1989;56:527–537. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

