Histone demethylase dUTX antagonizes JAK-STAT signaling to maintain proper gene expression and architecture of the Drosophila testis niche
- PMID: 23364332
- PMCID: PMC3583039
- DOI: 10.1242/dev.089433
Histone demethylase dUTX antagonizes JAK-STAT signaling to maintain proper gene expression and architecture of the Drosophila testis niche
Abstract
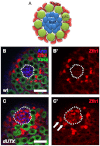
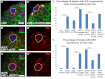
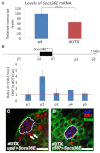
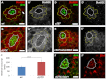
Adult stem cells reside in microenvironments called niches, where they are regulated by both extrinsic cues, such as signaling from neighboring cells, and intrinsic factors, such as chromatin structure. Here we report that in the Drosophila testis niche an H3K27me3-specific histone demethylase encoded by Ubiquitously transcribed tetratricopeptide repeat gene on the X chromosome (dUTX) maintains active transcription of the Suppressor of cytokine signaling at 36E (Socs36E) gene by removing the repressive H3K27me3 modification near its transcription start site. Socs36E encodes an inhibitor of the Janus kinase signal transducer and activator of transcription (JAK-STAT) signaling pathway. Whereas much is known about niche-to-stem cell signaling, such as the JAK-STAT signaling that is crucial for stem cell identity and activity, comparatively little is known about signaling from stem cells to the niche. Our results reveal that stem cells send feedback to niche cells to maintain the proper gene expression and architecture of the niche. We found that dUTX acts in cyst stem cells to maintain gene expression in hub cells through activating Socs36E transcription and preventing hyperactivation of JAK-STAT signaling. dUTX also acts in germline stem cells to maintain hub structure through regulating DE-Cadherin levels. Therefore, our findings provide new insights into how an epigenetic factor regulates crosstalk among different cell types within an endogenous stem cell niche, and shed light on the biological functions of a histone demethylase in vivo.
Figures
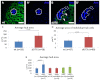
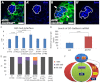
Similar articles
-
Competitiveness for the niche and mutual dependence of the germline and somatic stem cells in the Drosophila testis are regulated by the JAK/STAT signaling.J Cell Physiol. 2010 May;223(2):500-10. doi: 10.1002/jcp.22073. J Cell Physiol. 2010. PMID: 20143337 Free PMC article.
-
JAK-STAT signal inhibition regulates competition in the Drosophila testis stem cell niche.Science. 2009 Oct 2;326(5949):153-6. doi: 10.1126/science.1176817. Science. 2009. PMID: 19797664 Free PMC article.
-
A regulatory loop of JAK/STAT signalling and its downstream targets represses cell fate conversion and maintains male germline stem cell niche homeostasis.Cell Prolif. 2024 Oct;57(10):e13648. doi: 10.1111/cpr.13648. Epub 2024 Jul 10. Cell Prolif. 2024. PMID: 38987866 Free PMC article.
-
Hedgehog in the Drosophila testis niche: what does it do there?Protein Cell. 2013 Sep;4(9):650-5. doi: 10.1007/s13238-013-3040-y. Epub 2013 Jun 26. Protein Cell. 2013. PMID: 23807635 Free PMC article. Review.
-
Local signaling within stem cell niches: insights from Drosophila.Curr Opin Cell Biol. 2012 Apr;24(2):225-31. doi: 10.1016/j.ceb.2012.01.004. Epub 2012 Jan 30. Curr Opin Cell Biol. 2012. PMID: 22296770 Free PMC article. Review.
Cited by
-
Protecting and Diversifying the Germline.Genetics. 2018 Feb;208(2):435-471. doi: 10.1534/genetics.117.300208. Genetics. 2018. PMID: 29378808 Free PMC article. Review.
-
Heterochromatin Protein 1 (HP1) inhibits stem cell proliferation induced by ectopic activation of the Jak/STAT pathway in the Drosophila testis.Exp Cell Res. 2019 Apr 15;377(1-2):1-9. doi: 10.1016/j.yexcr.2019.02.024. Epub 2019 Feb 25. Exp Cell Res. 2019. PMID: 30817931 Free PMC article.
-
Drosophila Jak/STAT Signaling: Regulation and Relevance in Human Cancer and Metastasis.Int J Mol Sci. 2018 Dec 14;19(12):4056. doi: 10.3390/ijms19124056. Int J Mol Sci. 2018. PMID: 30558204 Free PMC article. Review.
-
Estrogen, through estrogen receptor 1, regulates histone modifications and chromatin remodeling during spermatogenesis in adult rats.Epigenetics. 2017;12(11):953-963. doi: 10.1080/15592294.2017.1382786. Epub 2017 Dec 18. Epigenetics. 2017. PMID: 28949791 Free PMC article.
-
A non-cell autonomous role of E(z) to prevent germ cells from turning on a somatic cell marker.Science. 2014 Mar 28;343(6178):1513-6. doi: 10.1126/science.1246514. Science. 2014. PMID: 24675960 Free PMC article.
References
-
- Agger K., Cloos P. A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A. E., Helin K. (2007). UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449, 731–734 - PubMed
-
- Bach E. A., Ekas L. A., Ayala-Camargo A., Flaherty M. S., Lee H., Perrimon N., Baeg G. H. (2007). GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr. Patterns 7, 323–331 - PubMed
-
- Becker P. B., Hörz W. (2002). ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71, 247–273 - PubMed
-
- Beebe K., Lee W. C., Micchelli C. A. (2010). JAK/STAT signaling coordinates stem cell proliferation and multilineage differentiation in the Drosophila intestinal stem cell lineage. Dev. Biol. 338, 28–37 - PubMed
-
- Beuchle D., Struhl G., Müller J. (2001). Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development 128, 993–1004 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

