The functional exchangeability of pk- and k-turns in RNA structure
- PMID: 23364423
- PMCID: PMC3672288
- DOI: 10.4161/rna.23673
The functional exchangeability of pk- and k-turns in RNA structure
Abstract
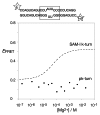
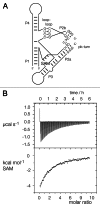
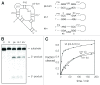
Ribonuclease P RNA requires a sharply kinked RNA helix to make a loop-receptor interaction that creates the binding site for the substrate. In some forms of the ribozyme, this is accomplished by a k-turn, while others have a different element called the pk-turn. The structure of the pk-turn in RNase P of Thermotoga maritima is globally very similar to a k-turn, but lacks all the standard features of that structure, including long-range hydrogen bonds between the two helical arms. We show here that in an isolated RNA duplex, the pk-turn fails to adopt a tightly kinked structure, but rather is a flexible element. This suggests that the tertiary contacts of RNase P assist its folding into the required kinked structure. We find that we can replace the k-turn of the SAM-I riboswitch with the pk-turn, such that the resulting RNA retains its ability to bind SAM, although with lower affinity. We also find that we can replace the pk-turn of T. maritima RNase P with a standard k-turn (in either orientation) with retention of ribozyme activity. Thus, although the pk-turn cannot intrinsically fold into the kinked structure, it can be induced to fold correctly in context. And the pk-turn and k-turns can substitute functionally for one another.
Keywords: RNA folding; RNA structure; RNase P; SAM-I riboswitch; kink turn.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
