The 'obligate diploid' Candida albicans forms mating-competent haploids
- PMID: 23364695
- PMCID: PMC3583542
- DOI: 10.1038/nature11865
The 'obligate diploid' Candida albicans forms mating-competent haploids
Erratum in
-
Corrigendum: The 'obligate diploid' Candida albicans forms mating-competent haploids.Nature. 2016 Feb 11;530(7589):242. doi: 10.1038/nature16134. Epub 2015 Nov 18. Nature. 2016. PMID: 26580011 Free PMC article. No abstract available.
Abstract
Candida albicans, the most prevalent human fungal pathogen, is considered to be an obligate diploid that carries recessive lethal mutations throughout the genome. Here we demonstrate that C. albicans has a viable haploid state that can be derived from diploid cells under in vitro and in vivo conditions, and that seems to arise through a concerted chromosome loss mechanism. Haploids undergo morphogenetic changes like those of diploids, including the yeast-hyphal transition, chlamydospore formation and a white-opaque switch that facilitates mating. Haploid opaque cells of opposite mating type mate efficiently to regenerate the diploid form, restoring heterozygosity and fitness. Homozygous diploids arise spontaneously by auto-diploidization, and both haploids and auto-diploids show a similar reduction in fitness, in vitro and in vivo, relative to heterozygous diploids, indicating that homozygous cell types are transient in mixed populations. Finally, we constructed stable haploid strains with multiple auxotrophies that will facilitate molecular and genetic analyses of this important pathogen.
Figures


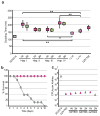
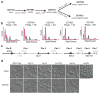
Comment in
-
Fungal biology: Multiple mating strategies.Nature. 2013 Feb 7;494(7435):45-6. doi: 10.1038/nature11945. Epub 2013 Jan 30. Nature. 2013. PMID: 23364693 No abstract available.
-
Fungal genetics: Candida gets a better half.Nat Rev Microbiol. 2013 Apr;11(4):225. doi: 10.1038/nrmicro2991. Epub 2013 Feb 18. Nat Rev Microbiol. 2013. PMID: 23419320 No abstract available.
References
Publication types
MeSH terms
Grants and funding
- R01 AI062427/AI/NIAID NIH HHS/United States
- T32 DE007288/DE/NIDCR NIH HHS/United States
- R21 AI081560/AI/NIAID NIH HHS/United States
- AI0624273/AI/NIAID NIH HHS/United States
- R01 AI0624273/AI/NIAID NIH HHS/United States
- P200A100100/PHS HHS/United States
- T32DE007288/DE/NIDCR NIH HHS/United States
- AI081560/AI/NIAID NIH HHS/United States
- R15 AI090633/AI/NIAID NIH HHS/United States
- R15-AI090633-01A1/AI/NIAID NIH HHS/United States
- R01 AI081704/AI/NIAID NIH HHS/United States
- R56 AI087401/AI/NIAID NIH HHS/United States
- AI081704/AI/NIAID NIH HHS/United States
- F32GM096536-02/GM/NIGMS NIH HHS/United States
- F32 GM096536/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

