Identification of a candidate therapeutic autophagy-inducing peptide
- PMID: 23364696
- PMCID: PMC3788641
- DOI: 10.1038/nature11866
Identification of a candidate therapeutic autophagy-inducing peptide
Abstract
The lysosomal degradation pathway of autophagy has a crucial role in defence against infection, neurodegenerative disorders, cancer and ageing. Accordingly, agents that induce autophagy may have broad therapeutic applications. One approach to developing such agents is to exploit autophagy manipulation strategies used by microbial virulence factors. Here we show that a peptide, Tat-beclin 1-derived from a region of the autophagy protein, beclin 1, which binds human immunodeficiency virus (HIV)-1 Nef-is a potent inducer of autophagy, and interacts with a newly identified negative regulator of autophagy, GAPR-1 (also called GLIPR2). Tat-beclin 1 decreases the accumulation of polyglutamine expansion protein aggregates and the replication of several pathogens (including HIV-1) in vitro, and reduces mortality in mice infected with chikungunya or West Nile virus. Thus, through the characterization of a domain of beclin 1 that interacts with HIV-1 Nef, we have developed an autophagy-inducing peptide that has potential efficacy in the treatment of human diseases.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
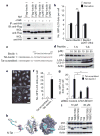
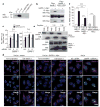


Comment in
-
Cell biology: Beneficial lessons from viruses.Nature. 2013 Feb 14;494(7436):181-2. doi: 10.1038/nature11947. Epub 2013 Jan 30. Nature. 2013. PMID: 23364685 No abstract available.
-
Peptides: activating autophagy.Nat Rev Drug Discov. 2013 Mar;12(3):190. doi: 10.1038/nrd3970. Nat Rev Drug Discov. 2013. PMID: 23449301 No abstract available.
-
A single pathway targets several health challenges of the elderly.Rejuvenation Res. 2014 Aug;17(4):382-4. doi: 10.1089/rej.2014.1585. Rejuvenation Res. 2014. PMID: 24953230
References
Publication types
MeSH terms
Substances
Grants and funding
- RC1 DK086502/DK/NIDDK NIH HHS/United States
- R01 NS063973/NS/NINDS NIH HHS/United States
- R0I NS077874/NS/NINDS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- 15153/CRUK_/Cancer Research UK/United Kingdom
- R01 NS084912/NS/NINDS NIH HHS/United States
- P30 CA142543/CA/NCI NIH HHS/United States
- R01 GM094575/GM/NIGMS NIH HHS/United States
- U54 AI057160/AI/NIAID NIH HHS/United States
- K08 AI099150/AI/NIAID NIH HHS/United States
- R01 GM079656/GM/NIGMS NIH HHS/United States
- U54 AI057156/AI/NIAID NIH HHS/United States
- R0I GM079656/GM/NIGMS NIH HHS/United States
- R01 GM066099/GM/NIGMS NIH HHS/United States
- R01 NS050199/NS/NINDS NIH HHS/United States
- U54AI057160/AI/NIAID NIH HHS/United States
- R0I GM066099/GM/NIGMS NIH HHS/United States
- T32 GM008297/GM/NIGMS NIH HHS/United States
- R01 NS077874/NS/NINDS NIH HHS/United States
- U54AI057156/AI/NIAID NIH HHS/United States
- R01 NS077111/NS/NINDS NIH HHS/United States
- R0I NS063973/NS/NINDS NIH HHS/United States
- R01 DK083756/DK/NIDDK NIH HHS/United States
- R0I DK086502/DK/NIDDK NIH HHS/United States
- R0I DK083756/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

