Structural analysis of the evolutionary origins of influenza virus hemagglutinin and other viral lectins
- PMID: 23365425
- PMCID: PMC3624229
- DOI: 10.1128/JVI.03476-12
Structural analysis of the evolutionary origins of influenza virus hemagglutinin and other viral lectins
Abstract
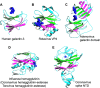
Influenza virus and other viruses use host cell surface sugars as receptors. Here we show that the sugar-binding domains in influenza virus hemagglutinin and other viral lectins share the same structural fold as human galectins (host lectins). Unlike the easily accessible sugar-binding sites in human galectins, the sugar-binding sites in viral lectins are hidden in cavities. We propose that these viral lectins originated from host lectins but have evolved to use hidden sugar-binding sites to evade host immune attacks.
Figures

References
-
- Wilson IA, Skehel JJ, Wiley DC. 1981. Structure of the hemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289:366–373 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

