Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues
- PMID: 23370273
- PMCID: PMC3783107
- DOI: 10.4161/pri.23807
Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues
Abstract
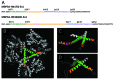
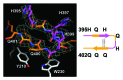
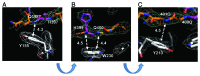
Huntington disease is an autosomal-dominant neurodegenerative disorder caused by a polyglutamine (polyQ) expansion (> 35Q) in the first exon (EX1) of huntingtin protein (Htt). mHtt protein is thought to adopt one or more toxic conformation(s) that are involved in pathogenic interactions in cells . However, the structure of mHtt is not known. Here, we present a near atomic resolution structure of mHtt36Q-EX1. To facilitate crystallization, three histidine residues (3H) were introduced within the Htt36Q stretch resulting in the sequence of Q 7HQHQHQ 27. The Htt36Q3H region adopts α-helix, loop, β-hairpin conformations. Furthermore, we observed interactions between the backbone of the Htt36Q3H β-strand with the aromatic residues mimicking putative-toxic interactions with other proteins. Our findings support previous predictions that the expanded mHtt-polyQ region adopts a β-sheet structure. Detailed structural information about mHtt improves our understanding of the pathogenic mechanisms in HD and other polyQ expansion disorders and may form the basis for rational design of small molecules that target toxic conformations of disease-causing proteins.
Keywords: Huntingtin; amyloid; crystallography; hairpin; polyglutamine; prion; sggregation; staxia; β-strand; β-turn.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
