PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root
- PMID: 23370718
- PMCID: PMC3561030
- DOI: 10.1104/pp.112.208678
PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root
Abstract
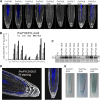
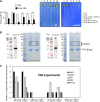
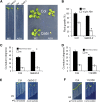
Abscisic acid (ABA) signaling plays a critical role in regulating root growth and root system architecture. ABA-mediated growth promotion and root tropic response under water stress are key responses for plant survival under limiting water conditions. In this work, we have explored the role of Arabidopsis (Arabidopsis thaliana) PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE (PYL)/REGULATORY COMPONENTS OF ABA RECEPTORS for root ABA signaling. As a result, we discovered that PYL8 plays a nonredundant role for the regulation of root ABA sensitivity. Unexpectedly, given the multigenic nature and partial functional redundancy observed in the PYR/PYL family, the single pyl8 mutant showed reduced sensitivity to ABA-mediated root growth inhibition. This effect was due to the lack of PYL8-mediated inhibition of several clade A phosphatases type 2C (PP2Cs), since PYL8 interacted in vivo with at least five PP2Cs, namely HYPERSENSITIVE TO ABA1 (HAB1), HAB2, ABA-INSENSITIVE1 (ABI1), ABI2, and PP2CA/ABA-HYPERSENSITIVE GERMINATION3 as revealed by tandem affinity purification and mass spectrometry proteomic approaches. We also discovered that PYR/PYL receptors and clade A PP2Cs are crucial for the hydrotropic response that takes place to guide root growth far from regions with low water potential. Thus, an ABA-hypersensitive pp2c quadruple mutant showed enhanced hydrotropism, whereas an ABA-insensitive sextuple pyr/pyl mutant showed reduced hydrotropic response, indicating that ABA-dependent inhibition of PP2Cs by PYR/PYLs is required for the proper perception of a moisture gradient.
Figures

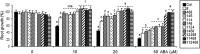
References
-
- Barrero JM, Rodriguez PL, Quesada V, Piqueras P, Ponce MR, Micol JL. (2006) Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ 29: 2000–2008 - PubMed
-
- Bensmihen S, To A, Lambert G, Kroj T, Giraudat J, Parcy F. (2004) Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett 561: 127–131 - PubMed
-
- Boudsocq M, Droillard MJ, Barbier-Brygoo H, Laurière C. (2007) Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol Biol 63: 491–503 - PubMed
-
- Bürckstümmer T, Bennett KL, Preradovic A, Schütze G, Hantschel O, Superti-Furga G, Bauch A. (2006) An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods 3: 1013–1019 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

