Does wheat genetically modified for disease resistance affect root-colonizing pseudomonads and arbuscular mycorrhizal fungi?
- PMID: 23372672
- PMCID: PMC3553117
- DOI: 10.1371/journal.pone.0053825
Does wheat genetically modified for disease resistance affect root-colonizing pseudomonads and arbuscular mycorrhizal fungi?
Abstract
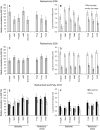
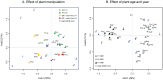
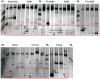
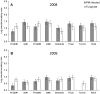
This study aimed to evaluate the impact of genetically modified (GM) wheat with introduced pm3b mildew resistance transgene, on two types of root-colonizing microorganisms, namely pseudomonads and arbuscular mycorrhizal fungi (AMF). Our investigations were carried out in field trials over three field seasons and at two locations. Serial dilution in selective King's B medium and microscopy were used to assess the abundance of cultivable pseudomonads and AMF, respectively. We developed a denaturing gradient gel electrophoresis (DGGE) method to characterize the diversity of the pqqC gene, which is involved in Pseudomonas phosphate solubilization. A major result was that in the first field season Pseudomonas abundances and diversity on roots of GM pm3b lines, but also on non-GM sister lines were different from those of the parental lines and conventional wheat cultivars. This indicates a strong effect of the procedures by which these plants were created, as GM and sister lines were generated via tissue cultures and propagated in the greenhouse. Moreover, Pseudomonas population sizes and DGGE profiles varied considerably between individual GM lines with different genomic locations of the pm3b transgene. At individual time points, differences in Pseudomonas and AMF accumulation between GM and control lines were detected, but they were not consistent and much less pronounced than differences detected between young and old plants, different conventional wheat cultivars or at different locations and field seasons. Thus, we conclude that impacts of GM wheat on plant-beneficial root-colonizing microorganisms are minor and not of ecological importance. The cultivation-independent pqqC-DGGE approach proved to be a useful tool for monitoring the dynamics of Pseudomonas populations in a wheat field and even sensitive enough for detecting population responses to altered plant physiology.
Conflict of interest statement
Figures
Similar articles
-
Competitive performance of transgenic wheat resistant to powdery mildew.PLoS One. 2011;6(11):e28091. doi: 10.1371/journal.pone.0028091. Epub 2011 Nov 23. PLoS One. 2011. PMID: 22132219 Free PMC article.
-
Interplay between wheat cultivars, biocontrol pseudomonads, and soil.Appl Environ Microbiol. 2010 Sep;76(18):6196-204. doi: 10.1128/AEM.00752-10. Epub 2010 Jul 30. Appl Environ Microbiol. 2010. PMID: 20675454 Free PMC article.
-
Molecular diversity and distribution of indigenous arbuscular mycorrhizal communities colonizing roots of two different winter cover crops in response to their root proliferation.J Microbiol. 2016 Feb;54(2):86-97. doi: 10.1007/s12275-016-5379-2. Epub 2016 Feb 2. J Microbiol. 2016. PMID: 26832664
-
Transgenic Pm3 multilines of wheat show increased powdery mildew resistance in the field.Plant Biotechnol J. 2012 May;10(4):398-409. doi: 10.1111/j.1467-7652.2011.00670.x. Epub 2011 Dec 18. Plant Biotechnol J. 2012. PMID: 22176579
-
Transgenic Pm3b wheat lines show resistance to powdery mildew in the field.Plant Biotechnol J. 2011 Oct;9(8):897-910. doi: 10.1111/j.1467-7652.2011.00603.x. Epub 2011 Mar 25. Plant Biotechnol J. 2011. PMID: 21438988
Cited by
-
The plant microbiome.Genome Biol. 2013 Jun 25;14(6):209. doi: 10.1186/gb-2013-14-6-209. Genome Biol. 2013. PMID: 23805896 Free PMC article. Review.
-
Plant-microbiome interactions for sustainable agriculture: a review.Physiol Mol Biol Plants. 2021 Jan;27(1):165-179. doi: 10.1007/s12298-021-00927-1. Epub 2021 Jan 30. Physiol Mol Biol Plants. 2021. PMID: 33627969 Free PMC article. Review.
-
Community Structure of Arbuscular Mycorrhizal Fungi in Rhizospheric Soil of a Transgenic High-Methionine Soybean and a Near Isogenic Variety.PLoS One. 2015 Dec 14;10(12):e0145001. doi: 10.1371/journal.pone.0145001. eCollection 2015. PLoS One. 2015. PMID: 26658560 Free PMC article.
-
Antimicrobial peptide expression in a wild tobacco plant reveals the limits of host-microbe-manipulations in the field.Elife. 2018 Apr 17;7:e28715. doi: 10.7554/eLife.28715. Elife. 2018. PMID: 29661271 Free PMC article.
-
Assessing Fungal Population in Soil Planted with Cry1Ac and CPTI Transgenic Cotton and Its Conventional Parental Line Using 18S and ITS rDNA Sequences over Four Seasons.Front Plant Sci. 2016 Jul 12;7:1023. doi: 10.3389/fpls.2016.01023. eCollection 2016. Front Plant Sci. 2016. PMID: 27462344 Free PMC article.
References
-
- Haas D, Défago G (2005) Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3: 307–319. - PubMed
-
- Haas D, Keel C (2003) Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu Rev Phytopathol 41: 117–153. - PubMed
-
- Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, et al. (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332: 1097–1100. - PubMed
-
- Goldstein AH (1986) Bacterial solubilization of mineralphosphates: historical perspective and future prospects. Am J Alternative Agr 1: 51–57.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

