Identification of novel biomarkers for sepsis prognosis via urinary proteomic analysis using iTRAQ labeling and 2D-LC-MS/MS
- PMID: 23372690
- PMCID: PMC3553154
- DOI: 10.1371/journal.pone.0054237
Identification of novel biomarkers for sepsis prognosis via urinary proteomic analysis using iTRAQ labeling and 2D-LC-MS/MS
Abstract
Objectives: Sepsis is the major cause of death for critically ill patients. Recent progress in proteomics permits a thorough characterization of the mechanisms associated with critical illness. The purpose of this study was to screen potential biomarkers for early prognostic assessment of patients with sepsis.
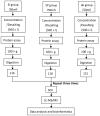
Methods: For the discovery stage, 30 sepsis patients with different prognoses were selected. Urinary proteins were identified using isobaric tags for relative and absolute quantitation (iTRAQ) coupled with LC-MS/MS. Mass spec instrument analysis were performed with Mascot software and the International Protein Index (IPI); bioinformatic analyses were used by the algorithm of set and the Gene Ontology (GO) Database. For the verification stage, the study involved another 54 sepsis-hospitalized patients, with equal numbers of patients in survivor and non-survivor groups based on 28-day survival. Differentially expressed proteins were verified by Western Blot.
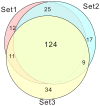
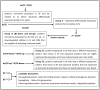
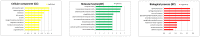
Results: A total of 232 unique proteins were identified. Proteins that were differentially expressed were further analyzed based on the pathophysiology of sepsis and biomathematics. For sepsis prognosis, five proteins were significantly up-regulated: selenium binding protein-1, heparan sulfate proteoglycan-2, alpha-1-B glycoprotein, haptoglobin, and lipocalin; two proteins were significantly down-regulated: lysosome-associated membrane proteins-1 and dipeptidyl peptidase-4. Based on gene ontology clustering, these proteins were associated with the biological processes of lipid homeostasis, cartilage development, iron ion transport, and certain metabolic processes. Urinary LAMP-1 was down-regulated, consistent with the Western Blot validation.
Conclusion: This study provides the proteomic analysis of urine to identify prognostic biomarkers of sepsis. The seven identified proteins provide insight into the mechanism of sepsis. Low urinary LAMP-1 levels may be useful for early prognostic assessment of sepsis.
Trial registration: ClinicalTrial.gov NCT01493492.
Conflict of interest statement
Figures
Similar articles
-
Identification of novel biomarkers for sepsis diagnosis via serum proteomic analysis using iTRAQ-2D-LC-MS/MS.J Clin Lab Anal. 2022 Jan;36(1):e24142. doi: 10.1002/jcla.24142. Epub 2021 Nov 26. J Clin Lab Anal. 2022. PMID: 34825737 Free PMC article.
-
Urinary proteomics analysis for sepsis biomarkers with iTRAQ labeling and two-dimensional liquid chromatography-tandem mass spectrometry.J Trauma Acute Care Surg. 2013 Mar;74(3):940-5. doi: 10.1097/TA.0b013e31828272c5. J Trauma Acute Care Surg. 2013. PMID: 23425763 Clinical Trial.
-
Identification and Verification of Two Novel Differentially Expressed Proteins from Non-neoplastic Mucosa and Colorectal Carcinoma Via iTRAQ Combined with Liquid Chromatography-Mass Spectrometry.Pathol Oncol Res. 2020 Apr;26(2):967-976. doi: 10.1007/s12253-019-00651-y. Epub 2019 Mar 29. Pathol Oncol Res. 2020. PMID: 30927204
-
Quantitative proteomics analysis to identify diffuse axonal injury biomarkers in rats using iTRAQ coupled LC-MS/MS.J Proteomics. 2016 Feb 5;133:93-99. doi: 10.1016/j.jprot.2015.12.014. Epub 2015 Dec 19. J Proteomics. 2016. PMID: 26710722 Review.
-
Discovery of urinary biomarkers.Mol Cell Proteomics. 2006 Oct;5(10):1760-71. doi: 10.1074/mcp.R600004-MCP200. Epub 2006 Jul 12. Mol Cell Proteomics. 2006. PMID: 16837576 Review.
Cited by
-
Identification of a novel sepsis prognosis model and analysis of possible drug application prospects: Based on scRNA-seq and RNA-seq data.Front Immunol. 2022 Oct 28;13:888891. doi: 10.3389/fimmu.2022.888891. eCollection 2022. Front Immunol. 2022. PMID: 36389695 Free PMC article.
-
Proteomics, physiological, and biochemical analysis of cross tolerance mechanisms in response to heat and water stresses in soybean.PLoS One. 2020 Jun 5;15(6):e0233905. doi: 10.1371/journal.pone.0233905. eCollection 2020. PLoS One. 2020. PMID: 32502194 Free PMC article.
-
Identification of novel biomarkers for sepsis diagnosis via serum proteomic analysis using iTRAQ-2D-LC-MS/MS.J Clin Lab Anal. 2022 Jan;36(1):e24142. doi: 10.1002/jcla.24142. Epub 2021 Nov 26. J Clin Lab Anal. 2022. PMID: 34825737 Free PMC article.
-
Sepsis: deriving biological meaning and clinical applications from high-dimensional data.Intensive Care Med Exp. 2021 May 7;9(1):27. doi: 10.1186/s40635-021-00383-x. Intensive Care Med Exp. 2021. PMID: 33961170 Free PMC article. Review.
-
Functional interaction of nicotinic acetylcholine receptors and Na+/K+ ATPase from Locusta migratoria manilensis (Meyen).Sci Rep. 2015 Mar 6;5:8849. doi: 10.1038/srep08849. Sci Rep. 2015. PMID: 25743085 Free PMC article.
References
-
- Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, et al. (2001) Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med 29: 1303–1310. - PubMed
-
- Gustot T (2011) Multiple organ failure in sepsis: prognosis and role of systemic inflammatory response. Curr Opin Crit Care 17: 153–159. - PubMed
-
- Vincent JL (2011) Acute kidney injury, acute lung injury and septic shock: how does mortality compare? Contrib Nephrol 174: 71–77. - PubMed
-
- Ricci Z, Ronco C (2009) Pathogenesis of acute kidney injury during sepsis. Curr Drug Targets 10: 1179–1183. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

