The transcription factor Runx2 is under circadian control in the suprachiasmatic nucleus and functions in the control of rhythmic behavior
- PMID: 23372705
- PMCID: PMC3555987
- DOI: 10.1371/journal.pone.0054317
The transcription factor Runx2 is under circadian control in the suprachiasmatic nucleus and functions in the control of rhythmic behavior
Abstract
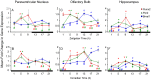
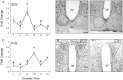
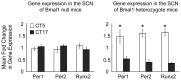
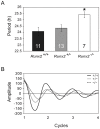
Runx2, a member of the family of runt-related transcription factors, is rhythmically expressed in bone and may be involved in circadian rhythms in bone homeostasis and osteogenesis. Runx2 is also expressed in the brain, but its function is unknown. We tested the hypothesis that in the brain, Runx2 may interact with clock-controlled genes to regulate circadian rhythms in behavior. First, we demonstrated diurnal and circadian rhythms in the expression of Runx2 in the mouse brain. Expression of Runx2 mRNA and protein mirrored that of the core clock genes, Period1 and Period2, in the suprachiasmatic nucleus (SCN), the paraventricular nucleus and the olfactory bulb. The rhythm of Runx2 expression was eliminated in the SCN of Bmal1(-/-) mice. Moreover, by crossbreeding mPer2(Luc) mice with Runx2(+/-) mice and recording bioluminescence rhythms, a significant lengthening of the period of rhythms was detected in cultured SCN of Runx2(-/-) animals compared to either Runx2(+/-) or Runx2(+/+) mice. Behavioral analyses of Runx2 mutant mice revealed that Runx2(+/-) animals displayed a significantly lengthened free-running period of running wheel activity compared to Runx2(+/+) littermates. Taken together, these findings provide evidence for clock gene-mediated rhythmic expression of Runx2, and its functional role in regulating circadian period at the level of the SCN and behavior.
Conflict of interest statement
Figures
Similar articles
-
Variations in Phase and Amplitude of Rhythmic Clock Gene Expression across Prefrontal Cortex, Hippocampus, Amygdala, and Hypothalamic Paraventricular and Suprachiasmatic Nuclei of Male and Female Rats.J Biol Rhythms. 2015 Oct;30(5):417-36. doi: 10.1177/0748730415598608. Epub 2015 Aug 13. J Biol Rhythms. 2015. PMID: 26271538 Free PMC article.
-
Circadian PER2::LUC rhythms in the olfactory bulb of freely moving mice depend on the suprachiasmatic nucleus but not on behaviour rhythms.Eur J Neurosci. 2015 Dec;42(12):3128-37. doi: 10.1111/ejn.13111. Eur J Neurosci. 2015. PMID: 26489367
-
Divergent roles of clock genes in retinal and suprachiasmatic nucleus circadian oscillators.PLoS One. 2012;7(6):e38985. doi: 10.1371/journal.pone.0038985. Epub 2012 Jun 11. PLoS One. 2012. PMID: 22701739 Free PMC article.
-
The Retinal Circadian Clock and Photoreceptor Viability.Adv Exp Med Biol. 2018;1074:345-350. doi: 10.1007/978-3-319-75402-4_42. Adv Exp Med Biol. 2018. PMID: 29721962 Free PMC article. Review.
-
Prokineticin 2 and circadian clock output.FEBS J. 2005 Nov;272(22):5703-9. doi: 10.1111/j.1742-4658.2005.04984.x. FEBS J. 2005. PMID: 16279936 Free PMC article. Review.
Cited by
-
The brain dynamics of linguistic computation.Front Psychol. 2015 Oct 13;6:1515. doi: 10.3389/fpsyg.2015.01515. eCollection 2015. Front Psychol. 2015. PMID: 26528201 Free PMC article.
-
Proteomic insights into circadian transcription regulation: novel E-box interactors revealed by proximity labeling.Genes Dev. 2024 Nov 27;38(21-24):1020-1032. doi: 10.1101/gad.351836.124. Genes Dev. 2024. PMID: 39562139 Free PMC article.
-
The tick tock of odontogenesis.Exp Cell Res. 2014 Jul 15;325(2):83-9. doi: 10.1016/j.yexcr.2014.02.007. Epub 2014 Feb 25. Exp Cell Res. 2014. PMID: 24582863 Free PMC article. Review.
-
BMAL1 Promotes Valvular Interstitial Cells' Osteogenic Differentiation through NF-κ B/AKT/MAPK Pathway.J Cardiovasc Dev Dis. 2023 Mar 6;10(3):110. doi: 10.3390/jcdd10030110. J Cardiovasc Dev Dis. 2023. PMID: 36975874 Free PMC article.
-
Multifactorial etiology of progressive supranuclear palsy (PSP): the genetic component.Acta Neuropathol. 2025 Jun 4;149(1):58. doi: 10.1007/s00401-025-02898-z. Acta Neuropathol. 2025. PMID: 40465013 Free PMC article. Review.
References
-
- Buijs RM, Kalsbeek A (2001) Hypothalamic integration of central and peripheral clocks. Nat Rev Neurosci 2: 521–526. - PubMed
-
- Buijs RM, van Eden CG, Goncharuk VD, Kalsbeek A (2003) The biological clock tunes the organs of the body: timing by hormones and the autonomic nervous system. J Endocrinol 177: 17–26. - PubMed
-
- Zhang EE, Kay SA (2010) Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol 11: 764–776. - PubMed
-
- Kawamoto T, Noshiro M, Sato F, Maemura K, Takeda N, et al. (2004) A novel autofeedback loop of Dec1 transcription involved in circadian rhythm regulation. Biochem Biophys Res Commun 313: 117–124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

