Structural and functional analysis of the DEAF-1 and BS69 MYND domains
- PMID: 23372760
- PMCID: PMC3555993
- DOI: 10.1371/journal.pone.0054715
Structural and functional analysis of the DEAF-1 and BS69 MYND domains
Abstract
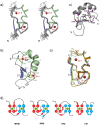
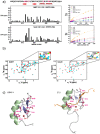
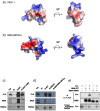
DEAF-1 is an important transcriptional regulator that is required for embryonic development and is linked to clinical depression and suicidal behavior in humans. It comprises various structural domains, including a SAND domain that mediates DNA binding and a MYND domain, a cysteine-rich module organized in a Cys(4)-Cys(2)-His-Cys (C4-C2HC) tandem zinc binding motif. DEAF-1 transcription regulation activity is mediated through interactions with cofactors such as NCoR and SMRT. Despite the important biological role of the DEAF-1 protein, little is known regarding the structure and binding properties of its MYND domain.Here, we report the solution structure, dynamics and ligand binding of the human DEAF-1 MYND domain encompassing residues 501-544 determined by NMR spectroscopy. The structure adopts a ββα fold that exhibits tandem zinc-binding sites with a cross-brace topology, similar to the MYND domains in AML1/ETO and other proteins. We show that the DEAF-1 MYND domain binds to peptides derived from SMRT and NCoR corepressors. The binding surface mapped by NMR titrations is similar to the one previously reported for AML1/ETO. The ligand binding and molecular functions of the related BS69 MYND domain were studied based on a homology model and mutational analysis. Interestingly, the interaction between BS69 and its binding partners (viral and cellular proteins) seems to require distinct charged residues flanking the predicted MYND domain fold, suggesting a different binding mode. Our findings demonstrate that the MYND domain is a conserved zinc binding fold that plays important roles in transcriptional regulation by mediating distinct molecular interactions with viral and cellular proteins.
Conflict of interest statement
Figures

Similar articles
-
BS69/ZMYND11 C-Terminal Domains Bind and Inhibit EBNA2.PLoS Pathog. 2016 Feb 4;12(2):e1005414. doi: 10.1371/journal.ppat.1005414. eCollection 2016 Feb. PLoS Pathog. 2016. PMID: 26845565 Free PMC article.
-
Structure and functional analysis of the MYND domain.J Mol Biol. 2006 Apr 28;358(2):498-508. doi: 10.1016/j.jmb.2006.01.087. Epub 2006 Feb 8. J Mol Biol. 2006. Retraction in: J Mol Biol. 2008 Mar 7;376(5):1523. doi: 10.1016/j.jmb.2008.01.071. PMID: 16527309 Retracted.
-
Increased association between Epstein-Barr virus EBNA2 from type 2 strains and the transcriptional repressor BS69 restricts EBNA2 activity.PLoS Pathog. 2019 Jul 8;15(7):e1007458. doi: 10.1371/journal.ppat.1007458. eCollection 2019 Jul. PLoS Pathog. 2019. PMID: 31283782 Free PMC article.
-
RING fingers and B-boxes: zinc-binding protein-protein interaction domains.Biochem Cell Biol. 1998;76(2-3):351-8. doi: 10.1139/bcb-76-2-3-351. Biochem Cell Biol. 1998. PMID: 9923704 Review.
-
Structure and Function of CW Domain Containing Proteins.Curr Protein Pept Sci. 2016;17(5):497-506. doi: 10.2174/1389203717666160125115130. Curr Protein Pept Sci. 2016. PMID: 26806410 Review.
Cited by
-
Mutations affecting the SAND domain of DEAF1 cause intellectual disability with severe speech impairment and behavioral problems.Am J Hum Genet. 2014 May 1;94(5):649-61. doi: 10.1016/j.ajhg.2014.03.013. Epub 2014 Apr 10. Am J Hum Genet. 2014. PMID: 24726472 Free PMC article.
-
Zn-regulated GTPase metalloprotein activator 1 modulates vertebrate zinc homeostasis.Cell. 2022 Jun 9;185(12):2148-2163.e27. doi: 10.1016/j.cell.2022.04.011. Epub 2022 May 17. Cell. 2022. PMID: 35584702 Free PMC article.
-
Refining analyses of copy number variation identifies specific genes associated with developmental delay.Nat Genet. 2014 Oct;46(10):1063-71. doi: 10.1038/ng.3092. Epub 2014 Sep 14. Nat Genet. 2014. PMID: 25217958 Free PMC article.
-
BS69/ZMYND11 C-Terminal Domains Bind and Inhibit EBNA2.PLoS Pathog. 2016 Feb 4;12(2):e1005414. doi: 10.1371/journal.ppat.1005414. eCollection 2016 Feb. PLoS Pathog. 2016. PMID: 26845565 Free PMC article.
-
DEAF1 binds unmethylated and variably spaced CpG dinucleotide motifs.PLoS One. 2014 Dec 22;9(12):e115908. doi: 10.1371/journal.pone.0115908. eCollection 2014. PLoS One. 2014. PMID: 25531106 Free PMC article.
References
-
- Michelson RJ, Collard MW, Ziemba AJ, Persinger J, Bartholomew B, et al. (1999) Nuclear DEAF-1-related (NUDR) Protein Contains a Novel DNA Binding Domain and Represses Transcription of the Heterogeneous Nuclear Ribonucleoprotein A2/B1 Promoter. JBC 274: 30510–30519. - PubMed
-
- Veraksa A, Kennison J, McGinnis W (2002) DEAF-1 Function Is Essential for the Early Embryonic Development of Drosophila. genesis 33: 67–76. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

