Atad3 function is essential for early post-implantation development in the mouse
- PMID: 23372768
- PMCID: PMC3556029
- DOI: 10.1371/journal.pone.0054799
Atad3 function is essential for early post-implantation development in the mouse
Abstract
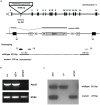
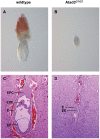
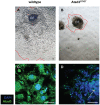
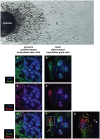
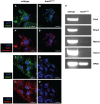
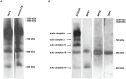
The mitochondrial AAA+-ATPase ATAD3 is implicated in the regulation of mitochondrial and ER dynamics and was shown to be necessary for larval development in Caenorhabditis elegans. In order to elucidate the relevance of ATAD3 for mammalian development, the phenotype of an Atad3 deficient mouse line was analyzed. Atad3 deficient embryos die around embryonic day E7.5 due to growth retardation and a defective development of the trophoblast lineage immediately after implantation into the uterus. This indicates an essential function of Atad3 for the progression of the first steps of post-implantation development at a time point when mitochondrial biogenesis and ATP production by oxidative phosphorylation are required. Therefore, murine Atad3 plays an important role in the biogenesis of mitochondria in trophoblast stem cells and in differentiating trophoblasts. At the biochemical level, we report here that ATAD3 is present in five native mitochondrial protein complexes of different sizes, indicating complex roles of the protein in mitochondrial architecture and function.
Conflict of interest statement
Figures
References
-
- Neuwald AF, Aravind L, Spouge JL, Koonin EV (1999) AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res 9: 27–43. - PubMed
-
- Langer T, Käser M, Klanner C, Leonhard K (2001) AAA proteases of mitochondria: quality control of membrane proteins and regulatory functions during mitochondrial biogenesis. Biochem Soc Trans 29: 431–6. - PubMed
-
- Hanson PI, Whiteheart SW (2005) AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol 6: 519–29. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

