The anaphase promoting complex contributes to the degradation of the S. cerevisiae telomerase recruitment subunit Est1p
- PMID: 23372810
- PMCID: PMC3555863
- DOI: 10.1371/journal.pone.0055055
The anaphase promoting complex contributes to the degradation of the S. cerevisiae telomerase recruitment subunit Est1p
Abstract
Telomerase is a multi-subunit enzyme that reverse transcribes telomere repeats onto the ends of linear eukaryotic chromosomes and is therefore critical for genome stability. S. cerevisiae telomerase activity is cell-cycle regulated; telomeres are not elongated during G1 phase. Previous work has shown that Est1 protein levels are low during G1 phase, preventing telomerase complex assembly. However, the pathway targeting Est1p for degradation remained uncharacterized. Here, we show that Est1p stability through the cell cycle mirrors that of Clb2p, a known target of the Anaphase Promoting Complex (APC). Indeed, Est1p is stabilized by mutations in both essential and non-essential components of the APC. Mutations of putative Destruction boxes (D-boxes), regions shown to be important for recognition of known APC substrates, stabilize Est1p, suggesting that Est1p is likely to be targeted for degradation directly by the APC. However, we do not detect degradation or ubiquitination of recombinant Est1p by the APC in vitro, suggesting either that the recombinant protein lacks necessary post-translational modification and/or conformation, or that the APC affects Est1p degradation by an indirect mechanism. Together, these studies shed light on the regulation of yeast telomerase assembly and demonstrate a new connection between telomere maintenance and cell cycle regulation pathways.
Conflict of interest statement
Figures

 . (C) Quantification of data shown in (B), as described in Materials and Methods. The calculated half-lives were averaged from independent biological replicates: αF (α-factor), n = 7; HU (hydroxyurea), n = 4; NOC (nocodazole), n = 4. Error bars are standard deviation from the mean. Both HU and NOC are statistically different from αF by two-tailed t-test (p-values 1.1×10−5 and 1.1×10−6, respectively) as denoted by *.
. (C) Quantification of data shown in (B), as described in Materials and Methods. The calculated half-lives were averaged from independent biological replicates: αF (α-factor), n = 7; HU (hydroxyurea), n = 4; NOC (nocodazole), n = 4. Error bars are standard deviation from the mean. Both HU and NOC are statistically different from αF by two-tailed t-test (p-values 1.1×10−5 and 1.1×10−6, respectively) as denoted by *.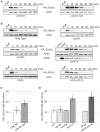

Similar articles
-
Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae.Nat Struct Mol Biol. 2006 Aug;13(8):720-8. doi: 10.1038/nsmb1125. Epub 2006 Jul 23. Nat Struct Mol Biol. 2006. PMID: 16862158
-
Anaphase promoting complex-dependent degradation of transcriptional repressors Nrm1 and Yhp1 in Saccharomyces cerevisiae.Mol Biol Cell. 2011 Jul 1;22(13):2175-84. doi: 10.1091/mbc.E11-01-0031. Epub 2011 May 11. Mol Biol Cell. 2011. PMID: 21562221 Free PMC article.
-
Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres.PLoS Genet. 2008 Oct;4(10):e1000236. doi: 10.1371/journal.pgen.1000236. Epub 2008 Oct 24. PLoS Genet. 2008. PMID: 18949040 Free PMC article.
-
Methods to measure ubiquitin-dependent proteolysis mediated by the anaphase-promoting complex.Methods. 2006 Jan;38(1):39-51. doi: 10.1016/j.ymeth.2005.07.005. Methods. 2006. PMID: 16343932 Review.
-
The anaphase-promoting complex (APC): the sum of its parts?Biochem Soc Trans. 2004 Nov;32(Pt 5):724-7. doi: 10.1042/BST0320724. Biochem Soc Trans. 2004. PMID: 15493998 Review.
Cited by
-
21st Century Genetics: Mass Spectrometry of Yeast Telomerase.Cold Spring Harb Symp Quant Biol. 2015;80:111-6. doi: 10.1101/sqb.2015.80.027656. Epub 2016 Jan 13. Cold Spring Harb Symp Quant Biol. 2015. PMID: 26763982 Free PMC article.
-
Identification of Saccharomyces cerevisiae Genes Whose Deletion Causes Synthetic Effects in Cells with Reduced Levels of the Nuclear Pif1 DNA Helicase.G3 (Bethesda). 2015 Oct 19;5(12):2913-8. doi: 10.1534/g3.115.021139. G3 (Bethesda). 2015. PMID: 26483010 Free PMC article.
-
Proteomic analysis defines the interactome of telomerase in the protozoan parasite, Trypanosoma brucei.Front Cell Dev Biol. 2023 Mar 16;11:1110423. doi: 10.3389/fcell.2023.1110423. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37009488 Free PMC article.
-
The anaphase promoting complex regulates yeast lifespan and rDNA stability by targeting Fob1 for degradation.Genetics. 2014 Mar;196(3):693-709. doi: 10.1534/genetics.113.158949. Epub 2013 Dec 20. Genetics. 2014. PMID: 24361936 Free PMC article.
-
Using Separation-of-Function Mutagenesis To Define the Full Spectrum of Activities Performed by the Est1 Telomerase Subunit in Vivo.Genetics. 2018 Jan;208(1):97-110. doi: 10.1534/genetics.117.300145. Epub 2017 Nov 29. Genetics. 2018. PMID: 29187507 Free PMC article.
References
-
- Wellinger RJ, Wolf AJ, Zakian VA (1993) Saccharomyces telomeres acquire single-strand TG1–3 tails late in S phase. Cell 72: 51–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

