Plasmodium falciparum sulfadoxine resistance is geographically and genetically clustered within the DR Congo
- PMID: 23372922
- PMCID: PMC3558697
- DOI: 10.1038/srep01165
Plasmodium falciparum sulfadoxine resistance is geographically and genetically clustered within the DR Congo
Abstract
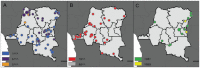
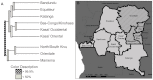
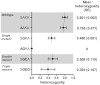
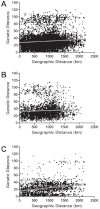
Understanding the spatial clustering of Plasmodium falciparum populations can assist efforts to contain drug-resistant parasites and maintain the efficacy of future drugs. We sequenced single nucleotide polymorphisms (SNPs) in the dihydropteroate synthase gene (dhps) associated with sulfadoxine resistance and 5 microsatellite loci flanking dhps in order to investigate the genetic backgrounds, genetic relatedness, and geographic clustering of falciparum parasites in the Democratic Republic of the Congo (DRC). Resistant haplotypes were clustered into subpopulations: one in the northeast DRC, and the other in the balance of the DRC. Network and clonal lineage analyses of the flanking microsatellites indicate that geographically-distinct mutant dhps haplotypes derive from separate lineages. The DRC is therefore a watershed for haplotypes associated with sulfadoxine resistance. Given the importance of central Africa as a corridor for the spread of antimalarial resistance, the identification of the mechanisms of this transit can inform future policies to contain drug-resistant parasite strains.
Figures

Similar articles
-
Different origin and dispersal of sulfadoxine-resistant Plasmodium falciparum haplotypes between Eastern Africa and Democratic Republic of Congo.Int J Antimicrob Agents. 2017 Apr;49(4):456-464. doi: 10.1016/j.ijantimicag.2016.12.007. Epub 2017 Feb 22. Int J Antimicrob Agents. 2017. PMID: 28237831
-
Independent lineages of highly sulfadoxine-resistant Plasmodium falciparum haplotypes, eastern Africa.Emerg Infect Dis. 2014 Jul;20(7):1140-8. doi: 10.3201/eid2007.131720. Emerg Infect Dis. 2014. PMID: 24960247 Free PMC article.
-
Multiple origins of Plasmodium falciparum dihydropteroate synthetase mutant alleles associated with sulfadoxine resistance in India.Antimicrob Agents Chemother. 2011 Jun;55(6):2813-7. doi: 10.1128/AAC.01151-10. Epub 2011 Mar 21. Antimicrob Agents Chemother. 2011. PMID: 21422213 Free PMC article.
-
Following the path of most resistance: dhps K540E dispersal in African Plasmodium falciparum.Trends Parasitol. 2010 Sep;26(9):447-56. doi: 10.1016/j.pt.2010.05.001. Epub 2010 Jun 17. Trends Parasitol. 2010. PMID: 20728060 Review.
-
Origins and spread of pfdhfr mutant alleles in Plasmodium falciparum.Acta Trop. 2010 Jun;114(3):166-70. doi: 10.1016/j.actatropica.2009.07.008. Epub 2009 Jul 14. Acta Trop. 2010. PMID: 19607799 Review.
Cited by
-
Genetic Evidence of Importation of Drug-Resistant Plasmodium falciparum to Guatemala from the Democratic Republic of the Congo.Emerg Infect Dis. 2014 Jun;20(6):932-40. doi: 10.3201/eid2006.131204. Emerg Infect Dis. 2014. PMID: 24856348 Free PMC article.
-
Malaria chemoprevention and drug resistance: a review of the literature and policy implications.Malar J. 2022 Mar 24;21(1):104. doi: 10.1186/s12936-022-04115-8. Malar J. 2022. PMID: 35331231 Free PMC article. Review.
-
Plasmodium falciparum genetic variation of var2csa in the Democratic Republic of the Congo.Malar J. 2018 Jan 24;17(1):46. doi: 10.1186/s12936-018-2193-9. Malar J. 2018. PMID: 29361940 Free PMC article.
-
Population Genetics and Molecular Epidemiology of Eukaryotes.Microbiol Spectr. 2018 Nov;6(6):10.1128/microbiolspec.ame-0002-2018. doi: 10.1128/microbiolspec.AME-0002-2018. Microbiol Spectr. 2018. PMID: 30387414 Free PMC article. Review.
-
A deep sequencing approach to estimate Plasmodium falciparum complexity of infection (COI) and explore apical membrane antigen 1 diversity.Malar J. 2017 Dec 16;16(1):490. doi: 10.1186/s12936-017-2137-9. Malar J. 2017. PMID: 29246158 Free PMC article.
References
-
- Wootton J. C. et al. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418, 320–323 (2002). - PubMed
-
- Roper C. et al. Intercontinental spread of pyrimethamine-resistant malaria. Science 305, 1124 (2004). - PubMed
-
- WHO. World Malaria Report 2011 (World Health Organization, 2012).
-
- Sibley C. H. et al. Pyrimethamine-sulfadoxine resistance in Plasmodium falciparum: what next? Trends Parasitol 17, 582–588 (2001). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

