Mechanisms of programmed DNA lesions and genomic instability in the immune system
- PMID: 23374339
- PMCID: PMC4382911
- DOI: 10.1016/j.cell.2013.01.007
Mechanisms of programmed DNA lesions and genomic instability in the immune system
Abstract
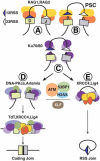
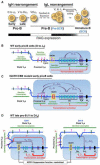
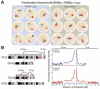
Chromosomal translocations involving antigen receptor loci are common in lymphoid malignancies. Translocations require DNA double-strand breaks (DSBs) at two chromosomal sites, their physical juxtaposition, and their fusion by end-joining. Ability of lymphocytes to generate diverse repertoires of antigen receptors and effector antibodies derives from programmed genomic alterations that produce DSBs. We discuss these lymphocyte-specific processes, with a focus on mechanisms that provide requisite DSB target specificity and mechanisms that suppress DSB translocation. We also discuss recent work that provides new insights into DSB repair pathways and the influences of three-dimensional genome organization on physiological processes and cancer genomes.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

