Genomic analysis of the regulatory elements and links with intrinsic DNA structural properties in the shrunken genome of Buchnera
- PMID: 23375088
- PMCID: PMC3571970
- DOI: 10.1186/1471-2164-14-73
Genomic analysis of the regulatory elements and links with intrinsic DNA structural properties in the shrunken genome of Buchnera
Abstract
Background: Buchnera aphidicola is an obligate symbiotic bacterium, associated with most of the aphididae, whose genome has drastically shrunk during intracellular evolution. Gene regulation in Buchnera has been a matter of controversy in recent years as the combination of genomic information with the experimental results has been contradictory, refuting or arguing in favour of a functional and responsive transcription regulation in Buchnera.The goal of this study was to describe the gene transcription regulation capabilities of Buchnera based on the inventory of cis- and trans-regulators encoded in the genomes of five strains from different aphids (Acyrthosiphon pisum, Schizaphis graminum, Baizongia pistacea, Cinara cedri and Cinara tujafilina), as well as on the characterisation of some intrinsic structural properties of the DNA molecule in these bacteria.
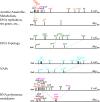
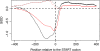
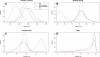
Results: Interaction graph analysis shows that gene neighbourhoods are conserved between E. coli and Buchnera in structures called transcriptons, interactons and metabolons, indicating that selective pressures have acted on the evolution of transcriptional, protein-protein interaction and metabolic networks in Buchnera. The transcriptional regulatory network in Buchnera is composed of a few general DNA-topological regulators (Nucleoid Associated Proteins and topoisomerases), with the quasi-absence of any specific ones (except for multifunctional enzymes with a known gene expression regulatory role in Escherichia coli, such as AlaS, PepA and BolA, and the uncharacterized hypothetical regulators YchA and YrbA). The relative positioning of regulatory genes along the chromosome of Buchnera seems to have conserved its ancestral state, despite the genome erosion. Sigma-70 promoters with canonical thermodynamic sequence profiles were detected upstream of about 94% of the CDS of Buchnera in the different aphids. Based on Stress-Induced Duplex Destabilization (SIDD) measurements, unstable σ70 promoters were found specifically associated with the regulator and transporter genes.
Conclusions: This genomic analysis provides supporting evidence of a selection of functional regulatory structures and it has enabled us to propose hypotheses concerning possible links between these regulatory elements and the DNA-topology (i.e., supercoiling, curvature, flexibility and base-pair stability) in the regulation of gene expression in the shrunken genome of Buchnera.
Figures
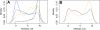
Similar articles
-
New clues about the evolutionary history of metabolic losses in bacterial endosymbionts, provided by the genome of Buchnera aphidicola from the aphid Cinara tujafilina.Appl Environ Microbiol. 2011 Jul;77(13):4446-54. doi: 10.1128/AEM.00141-11. Epub 2011 May 13. Appl Environ Microbiol. 2011. PMID: 21571878 Free PMC article.
-
A genomic reappraisal of symbiotic function in the aphid/Buchnera symbiosis: reduced transporter sets and variable membrane organisations.PLoS One. 2011;6(12):e29096. doi: 10.1371/journal.pone.0029096. Epub 2011 Dec 27. PLoS One. 2011. PMID: 22229056 Free PMC article.
-
Reinventing the Wheel and Making It Round Again: Evolutionary Convergence in Buchnera-Serratia Symbiotic Consortia between the Distantly Related Lachninae Aphids Tuberolachnus salignus and Cinara cedri.Genome Biol Evol. 2016 May 22;8(5):1440-58. doi: 10.1093/gbe/evw085. Genome Biol Evol. 2016. PMID: 27190007 Free PMC article.
-
Genomic revelations of a mutualism: the pea aphid and its obligate bacterial symbiont.Cell Mol Life Sci. 2011 Apr;68(8):1297-309. doi: 10.1007/s00018-011-0645-2. Epub 2011 Mar 10. Cell Mol Life Sci. 2011. PMID: 21390549 Free PMC article. Review.
-
Genomic identification of regulatory elements by evolutionary sequence comparison and functional analysis.Adv Genet. 2008;61:269-93. doi: 10.1016/S0065-2660(07)00010-7. Adv Genet. 2008. PMID: 18282510 Free PMC article. Review.
Cited by
-
Spatial and morphological reorganization of endosymbiosis during metamorphosis accommodates adult metabolic requirements in a weevil.Proc Natl Acad Sci U S A. 2020 Aug 11;117(32):19347-19358. doi: 10.1073/pnas.2007151117. Epub 2020 Jul 28. Proc Natl Acad Sci U S A. 2020. PMID: 32723830 Free PMC article.
-
Transcription Factors Exhibit Differential Conservation in Bacteria with Reduced Genomes.PLoS One. 2016 Jan 14;11(1):e0146901. doi: 10.1371/journal.pone.0146901. eCollection 2016. PLoS One. 2016. PMID: 26766575 Free PMC article.
-
Role of the Discriminator Sequence in the Supercoiling Sensitivity of Bacterial Promoters.mSystems. 2021 Aug 31;6(4):e0097821. doi: 10.1128/mSystems.00978-21. Epub 2021 Aug 24. mSystems. 2021. PMID: 34427530 Free PMC article.
-
A complete DNA repair system assembled by two endosymbionts restores heat tolerance of the insect host.Proc Natl Acad Sci U S A. 2024 Dec 17;121(51):e2415651121. doi: 10.1073/pnas.2415651121. Epub 2024 Dec 10. Proc Natl Acad Sci U S A. 2024. PMID: 39656210 Free PMC article.
-
Systematic discovery of uncharacterized transcription factors in Escherichia coli K-12 MG1655.Nucleic Acids Res. 2018 Nov 16;46(20):10682-10696. doi: 10.1093/nar/gky752. Nucleic Acids Res. 2018. PMID: 30137486 Free PMC article.
References
-
- Brinza L, Viñuelas J, Cottret L, Calevro F, Rahbé Y, Febvay G, Duport G, Colella S, Rabatel A, Gautier C. et al.Systemic analysis of the symbiotic function of Buchnera aphidicola, the primary endosymbiont of the pea aphid Acyrthosiphon pisum. C R Biol. 2009;332(11):1034–1049. doi: 10.1016/j.crvi.2009.09.007. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

