Intra-platform comparison of 25-mer and 60-mer oligonucleotide Nimblegen DNA microarrays
- PMID: 23375116
- PMCID: PMC3608165
- DOI: 10.1186/1756-0500-6-43
Intra-platform comparison of 25-mer and 60-mer oligonucleotide Nimblegen DNA microarrays
Abstract
Background: We performed a Nimblegen intra-platform microarray comparison by assessing two categories of flax target probes (short 25-mers oligonucleotides and long 60-mers oligonucleotides) in identical conditions of target production, design, labelling, hybridization, image analyses, and data filtering. We compared technical parameters of array hybridizations, precision and accuracy as well as specific gene expression profiles.
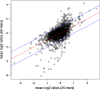
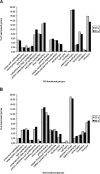
Results: Comparison of the hybridization quality, precision and accuracy of expression measurements, as well as an interpretation of differential gene expression in flax tissues were performed. Both array types yielded reproducible, accurate and comparable data that are coherent for expression measurements and identification of differentially expressed genes. 60-mers arrays gave higher hybridization efficiencies and therefore were more sensitive allowing the detection of a higher number of unigenes involved in the same biological process and/or belonging to the same multigene family.
Conclusion: The two flax arrays provide a good resolution of expressed functions; however the 60-mers arrays are more sensitive and provide a more in-depth coverage of candidate genes potentially involved in different biological processes.
Figures





References
-
- Dobbin KK, Kawasaki ES, Petersen DW, Simon RM. Characterizing dye bias in microarray experiments. Bioinformatics. 2005;17:17. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

