Analyses of the effects of all ubiquitin point mutants on yeast growth rate
- PMID: 23376099
- PMCID: PMC3615125
- DOI: 10.1016/j.jmb.2013.01.032
Analyses of the effects of all ubiquitin point mutants on yeast growth rate
Abstract
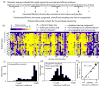
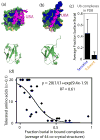
The amino acid sequence of a protein governs its function. We used bulk competition and focused deep sequencing to investigate the effects of all ubiquitin point mutants on yeast growth rate. Many aspects of ubiquitin function have been carefully studied, which enabled interpretation of our growth analyses in light of a rich structural, biophysical and biochemical knowledge base. In one highly sensitive cluster on the surface of ubiquitin, almost every amino acid substitution caused growth defects. In contrast, the opposite face tolerated virtually all possible substitutions. Surface locations between these two faces exhibited intermediate mutational tolerance. The sensitive face corresponds to the known interface for many binding partners. Across all surface positions, we observe a strong correlation between burial at structurally characterized interfaces and the number of amino acid substitutions compatible with robust growth. This result indicates that binding is a dominant determinant of ubiquitin function. In the solvent-inaccessible core of ubiquitin, all positions tolerated a limited number of substitutions, with hydrophobic amino acids especially interchangeable. Some mutations null for yeast growth were previously shown to populate folded conformations indicating that, for these mutants, subtle changes to conformation caused functional defects. The most sensitive region to mutation within the core was located near the C-terminus that is a focal binding site for many critical binding partners. These results indicate that core mutations may frequently cause functional defects through subtle disturbances to structure or dynamics.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures




Comment in
-
Alanine scan of core positions in ubiquitin reveals links between dynamics, stability, and function.J Mol Biol. 2014 Apr 3;426(7):1377-89. doi: 10.1016/j.jmb.2013.10.042. Epub 2013 Dec 19. J Mol Biol. 2014. PMID: 24361330 Free PMC article.
References
-
- Loeb DD, Swanstrom R, Everitt L, Manchester M, Stamper SE, Hutchison CA., 3rd Complete mutagenesis of the HIV-1 protease. Nature. 1989;340:397–400. - PubMed
-
- Rennell D, Bouvier SE, Hardy LW, Poteete AR. Systematic mutation of bacteriophage T4 lysozyme. J Mol Biol. 1991;222:67–88. - PubMed
-
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

