Integrated metabolite and gene expression profiles identify lipid biomarkers associated with progression of hepatocellular carcinoma and patient outcomes
- PMID: 23376425
- PMCID: PMC3633738
- DOI: 10.1053/j.gastro.2013.01.054
Integrated metabolite and gene expression profiles identify lipid biomarkers associated with progression of hepatocellular carcinoma and patient outcomes
Abstract
Background & aims: We combined gene expression and metabolic profiling analyses to identify factors associated with outcomes of patients with hepatocellular carcinoma (HCC).
Methods: We compared metabolic and gene expression patterns between paired tumor and nontumor tissues from 30 patients with HCC, and validated the results using samples from 356 patients with HCC. A total of 469 metabolites were measured using liquid chromatography/mass spectrometry and gas chromatography/mass spectrometry. Metabolic and genomic data were integrated, and Kaplan-Meier and Cox proportional hazards analyses were used to associate specific patterns with patient outcomes. Associated factors were evaluated for their effects on cancer cells in vitro and tumor formation in nude mice.
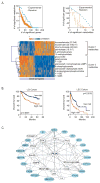
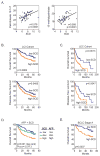
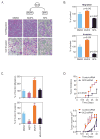
Results: We identified 28 metabolites and 169 genes associated with aggressive HCC. Lipid metabolites of stearoyl-CoA-desaturase (SCD) activity were associated with aberrant palmitate signaling in aggressive HCC samples. Expression of gene products associated with these metabolites, including SCD, were associated independently with survival times and tumor recurrence in the test and validation sets. Combined expression of SCD and α-fetoprotein were associated with outcomes of patients with early-stage HCC. Levels of monounsaturated palmitic acid, the product of SCD activity, were increased in aggressive HCCs; monounsaturated palmitic acid increased migration and invasion of cultured HCC cells and colony formation by HCC cells. HCC cells that expressed small interfering RNA against SCD had decreased cell migration and colony formation in culture and reduced tumorigenicity in mice.
Conclusions: By using a combination of gene expression and metabolic profile analysis, we identified a lipogenic network that involves SCD and palmitate signaling and was associated with HCC progression and patient outcomes. The microarray platform and data have been submitted to the Gene Expression Omnibus public database at NCBI following MIAME guidelines. Accession numbers: GPL4700 (platform), and GSE6857 (samples).
Copyright © 2013 AGA Institute. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Nielsen J, Oliver S. The next wave in metabolome analysis. Trends Biotechnol. 2005;23:544–546. - PubMed
-
- Heinemann M, Zenobi R. Single cell metabolomics. Curr Opin Biotechnol. 2011;22:26–31. - PubMed
-
- Budhu A, Forgues M, Ye QH, et al. Prediction of venous metastases, recurrence and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10:99–111. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

