Fusion genes and their discovery using high throughput sequencing
- PMID: 23376639
- PMCID: PMC3675181
- DOI: 10.1016/j.canlet.2013.01.011
Fusion genes and their discovery using high throughput sequencing
Abstract
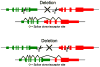
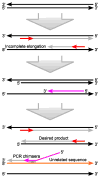
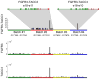
Fusion genes are hybrid genes that combine parts of two or more original genes. They can form as a result of chromosomal rearrangements or abnormal transcription, and have been shown to act as drivers of malignant transformation and progression in many human cancers. The biological significance of fusion genes together with their specificity to cancer cells has made them into excellent targets for molecular therapy. Fusion genes are also used as diagnostic and prognostic markers to confirm cancer diagnosis and monitor response to molecular therapies. High-throughput sequencing has enabled the systematic discovery of fusion genes in a wide variety of cancer types. In this review, we describe the history of fusion genes in cancer and the ways in which fusion genes form and affect cellular function. We also describe computational methodologies for detecting fusion genes from high-throughput sequencing experiments, and the most common sources of error that lead to false discovery of fusion genes.
Keywords: Cancer; Fusion gene; High throughput sequencing.
Copyright © 2013 Elsevier Ireland Ltd. All rights reserved.
Figures



References
-
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nature Reviews Cancer. 2007;7:233–245. - PubMed
-
- Nowell PC, Hungerford DA. A minute chromosome in human chronic granulocytic leukemia. Science. 1960;142:1497. - PubMed
-
- Rowley JD. A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. - PubMed
-
- Shtivelman E, Lifshitz B, Gale RP, Canaani E. Fused transcript of abl and bcr genes in chronic myelogenous leukaemia. Nature. 1985;315:550–554. - PubMed
-
- Manolov G, Manolova Y. Marker band in one chromosome 14 from Burkitt lymphomas. Nature. 1972;237:33–34. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

