TMEM59 defines a novel ATG16L1-binding motif that promotes local activation of LC3
- PMID: 23376921
- PMCID: PMC3579146
- DOI: 10.1038/emboj.2013.8
TMEM59 defines a novel ATG16L1-binding motif that promotes local activation of LC3
Abstract
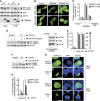
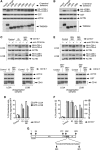
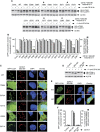
Selective autophagy underlies many of the important physiological roles that autophagy plays in multicellular organisms, but the mechanisms involved in cargo selection are poorly understood. Here we describe a molecular mechanism that can target conventional endosomes for autophagic degradation. We show that the human transmembrane protein TMEM59 contains a minimal 19-amino-acid peptide in its intracellular domain that promotes LC3 labelling and lysosomal targeting of its own endosomal compartment. Interestingly, this peptide defines a novel protein motif that mediates interaction with the WD-repeat domain of ATG16L1, thus providing a mechanistic basis for the activity. The motif is represented with the same ATG16L1-binding ability in other molecules, suggesting a more general relevance. We propose that this motif may play an important role in targeting specific membranous compartments for autophagic degradation, and therefore it may facilitate the search for adaptor proteins that promote selective autophagy by engaging ATG16L1. Endogenous TMEM59 interacts with ATG16L1 and mediates autophagy in response to Staphylococcus aureus infection.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






Similar articles
-
The T300A Crohn's disease risk polymorphism impairs function of the WD40 domain of ATG16L1.Nat Commun. 2016 Jun 8;7:11821. doi: 10.1038/ncomms11821. Nat Commun. 2016. PMID: 27273576 Free PMC article.
-
ATG16L1 WD40 domain-dependent IL10R (interleukin 10 receptor) signaling is insensitive to the T300A Crohn disease risk polymorphism.Autophagy. 2022 Dec;18(12):3023-3030. doi: 10.1080/15548627.2022.2054241. Epub 2022 Mar 29. Autophagy. 2022. PMID: 35311452 Free PMC article.
-
Missing WD40 Repeats in ATG16L1 Delays Canonical Autophagy and Inhibits Noncanonical Autophagy.Int J Mol Sci. 2024 Apr 19;25(8):4493. doi: 10.3390/ijms25084493. Int J Mol Sci. 2024. PMID: 38674078 Free PMC article.
-
Chaperone-mediated autophagy and endosomal microautophagy: Joint by a chaperone.J Biol Chem. 2018 Apr 13;293(15):5414-5424. doi: 10.1074/jbc.R117.818237. Epub 2017 Dec 15. J Biol Chem. 2018. PMID: 29247007 Free PMC article. Review.
-
Molecules and their functions in autophagy.Exp Mol Med. 2012 Feb 29;44(2):73-80. doi: 10.3858/emm.2012.44.2.029. Exp Mol Med. 2012. PMID: 22257882 Free PMC article. Review.
Cited by
-
Dcf1 Affects Memory and Anxiety by Regulating NMDA and AMPA Receptors.Neurochem Res. 2019 Nov;44(11):2499-2505. doi: 10.1007/s11064-019-02866-6. Epub 2019 Sep 17. Neurochem Res. 2019. PMID: 31531752
-
Genetic Analysis of Six Transmembrane Protein Family Genes in Parkinson's Disease in a Large Chinese Cohort.Front Aging Neurosci. 2022 Jul 4;14:889057. doi: 10.3389/fnagi.2022.889057. eCollection 2022. Front Aging Neurosci. 2022. PMID: 35860667 Free PMC article.
-
A transmembrane protein family gene signature for overall survival prediction in osteosarcoma.Front Genet. 2022 Aug 5;13:937300. doi: 10.3389/fgene.2022.937300. eCollection 2022. Front Genet. 2022. PMID: 35991561 Free PMC article.
-
Dcf1 Triggers Dendritic Spine Formation and Facilitates Memory Acquisition.Mol Neurobiol. 2018 Jan;55(1):763-775. doi: 10.1007/s12035-016-0349-6. Epub 2017 Jan 5. Mol Neurobiol. 2018. PMID: 28058580
-
An unconventional autophagic pathway that inhibits ATP secretion during apoptotic cell death.Nat Commun. 2025 Apr 10;16(1):3409. doi: 10.1038/s41467-025-58619-3. Nat Commun. 2025. PMID: 40210636 Free PMC article.
References
-
- Alcalá S, Klee M, Fernandez J, Fleischer A, Pimentel-Muiños FX (2008) A high-throughput screening for mammalian cell death effectors identifies the mitochondrial phosphate carrier as a regulator of cytochrome c release. Oncogene 27: 44–54 - PubMed
-
- Bonifacino JS, Traub LM (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem 72: 395–447 - PubMed
-
- Cadwell K, Stappenbeck TS, Virgin HW (2009) Role of autophagy and autophagy genes in inflammatory bowel disease. Curr Top Microbiol Immunol 335: 141–167 - PubMed
-
- Campoy E, Colombo MI (2009) Autophagy in intracellular bacterial infection. Biochim Biophys Acta 1793: 1465–1477 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

