Transcribed pseudogene ψPPM1K generates endogenous siRNA to suppress oncogenic cell growth in hepatocellular carcinoma
- PMID: 23376929
- PMCID: PMC3616710
- DOI: 10.1093/nar/gkt047
Transcribed pseudogene ψPPM1K generates endogenous siRNA to suppress oncogenic cell growth in hepatocellular carcinoma
Abstract
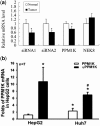
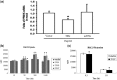
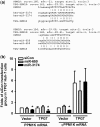
Pseudogenes, especially those that are transcribed, may not be mere genomic fossils, but their biological significance remains unclear. Postulating that in the human genome, as in animal models, pseudogenes may function as gene regulators through generation of endo-siRNAs (esiRNAs), antisense RNAs or RNA decoys, we performed bioinformatic and subsequent experimental tests to explore esiRNA-mediated mechanisms of pseudogene involvement in oncogenesis. A genome-wide survey revealed a partial retrotranscript pseudogene ψPPM1K containing inverted repeats capable of folding into hairpin structures that can be processed into two esiRNAs; these esiRNAs potentially target many cellular genes, including NEK8. In 41 paired surgical specimens, we found significantly reduced expression of two predicted ψPPM1K-specific esiRNAs, and the cognate gene PPM1K, in hepatocellular carcinoma compared with matched non-tumour tissues, whereas the expression of target gene NEK8 was increased in tumours. Additionally, NEK8 and PPM1K were downregulated in stably transfected ψPPM1K-overexpressing cells, but not in cells transfected with an esiRNA1-deletion mutant of ψPPM1K. Furthermore, expression of NEK8 in ψPPM1K-transfected cells demonstrated that NEK8 can counteract the growth inhibitory effects of ψPPM1K. These findings indicate that a transcribed pseudogene can exert tumour-suppressor activity independent of its parental gene by generation of esiRNAs that regulate human cell growth.
Figures







Similar articles
-
Pseudogene-derived endogenous siRNAs and their function.Methods Mol Biol. 2014;1167:227-39. doi: 10.1007/978-1-4939-0835-6_15. Methods Mol Biol. 2014. PMID: 24823781 Review.
-
Pseudogene INTS6P1 regulates its cognate gene INTS6 through competitive binding of miR-17-5p in hepatocellular carcinoma.Oncotarget. 2015 Mar 20;6(8):5666-77. doi: 10.18632/oncotarget.3290. Oncotarget. 2015. PMID: 25686840 Free PMC article.
-
Pseudogene AKR1B10P1 enhances tumorigenicity and regulates epithelial-mesenchymal transition in hepatocellular carcinoma via stabilizing SOX4.J Cell Mol Med. 2020 Oct;24(20):11779-11790. doi: 10.1111/jcmm.15790. Epub 2020 Sep 13. J Cell Mol Med. 2020. PMID: 32924268 Free PMC article.
-
Integrated analysis of pseudogene RP11-564D11.3 expression and its potential roles in hepatocellular carcinoma.Epigenomics. 2019 Feb;11(3):267-280. doi: 10.2217/epi-2018-0152. Epub 2018 Oct 26. Epigenomics. 2019. PMID: 30362374
-
Pseudogenes: pseudo-functional or key regulators in health and disease?RNA. 2011 May;17(5):792-8. doi: 10.1261/rna.2658311. Epub 2011 Mar 11. RNA. 2011. PMID: 21398401 Free PMC article. Review.
Cited by
-
Pseudogene BMI1P1 expression as a novel predictor for acute myeloid leukemia development and prognosis.Oncotarget. 2016 Jul 26;7(30):47376-47386. doi: 10.18632/oncotarget.10156. Oncotarget. 2016. PMID: 27329719 Free PMC article.
-
Construction of survival-related co-expression modules and identification of potential prognostic biomarkers of osteosarcoma using WGCNA.Ann Transl Med. 2022 Mar;10(6):296. doi: 10.21037/atm-22-399. Ann Transl Med. 2022. PMID: 35434042 Free PMC article.
-
Role of Pseudogenes in Tumorigenesis.Cancers (Basel). 2018 Aug 1;10(8):256. doi: 10.3390/cancers10080256. Cancers (Basel). 2018. PMID: 30071685 Free PMC article. Review.
-
Identification of Potential Prostate Cancer-Related Pseudogenes Based on Competitive Endogenous RNA Network Hypothesis.Med Sci Monit. 2018 Jun 20;24:4213-4239. doi: 10.12659/MSM.910886. Med Sci Monit. 2018. PMID: 29923546 Free PMC article.
-
Structural basis of endo-siRNA processing by Drosophila Dicer-2 and Loqs-PD.Nucleic Acids Res. 2025 Feb 8;53(4):gkaf102. doi: 10.1093/nar/gkaf102. Nucleic Acids Res. 2025. PMID: 39988314 Free PMC article.
References
-
- Mighell AJ, Smith NR, Robinson PA, Markham AF. Vertebrate pseudogenes. FEBS Lett. 2000;468:109–114. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

