Eukaryotic TPP riboswitch regulation of alternative splicing involving long-distance base pairing
- PMID: 23376932
- PMCID: PMC3597705
- DOI: 10.1093/nar/gkt057
Eukaryotic TPP riboswitch regulation of alternative splicing involving long-distance base pairing
Abstract
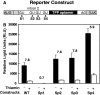
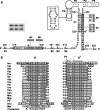
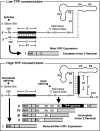
Thiamin pyrophosphate (TPP) riboswitches are found in organisms from all three domains of life. Examples in bacteria commonly repress gene expression by terminating transcription or by blocking ribosome binding, whereas most eukaryotic TPP riboswitches are predicted to regulate gene expression by modulating RNA splicing. Given the widespread distribution of eukaryotic TPP riboswitches and the diversity of their locations in precursor messenger RNAs (pre-mRNAs), we sought to examine the mechanism of alternative splicing regulation by a fungal TPP riboswitch from Neurospora crassa, which is mostly located in a large intron separating protein-coding exons. Our data reveal that this riboswitch uses a long-distance (∼530-nt separation) base-pairing interaction to regulate alternative splicing. Specifically, a portion of the TPP-binding aptamer can form a base-paired structure with a conserved sequence element (α) located near a 5' splice site, which greatly increases use of this 5' splice site and promotes gene expression. Comparative sequence analyses indicate that many fungal species carry a TPP riboswitch with similar intron architecture, and therefore the homologous genes in these fungi are likely to use the same mechanism. Our findings expand the scope of genetic control mechanisms relying on long-range RNA interactions to include riboswitches.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

