Hyper conserved elements in vertebrate mRNA 3'-UTRs reveal a translational network of RNA-binding proteins controlled by HuR
- PMID: 23376935
- PMCID: PMC3597683
- DOI: 10.1093/nar/gkt017
Hyper conserved elements in vertebrate mRNA 3'-UTRs reveal a translational network of RNA-binding proteins controlled by HuR
Abstract
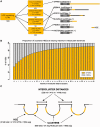
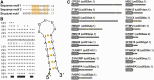
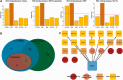
Little is known regarding the post-transcriptional networks that control gene expression in eukaryotes. Additionally, we still need to understand how these networks evolve, and the relative role played in them by their sequence-dependent regulatory factors, non-coding RNAs (ncRNAs) and RNA-binding proteins (RBPs). Here, we used an approach that relied on both phylogenetic sequence sharing and conservation in the whole mapped 3'-untranslated regions (3'-UTRs) of vertebrate species to gain knowledge on core post-transcriptional networks. The identified human hyper conserved elements (HCEs) were predicted to be preferred binding sites for RBPs and not for ncRNAs, namely microRNAs and long ncRNAs. We found that the HCE map identified a well-known network that post-transcriptionally regulates histone mRNAs. We were then able to discover and experimentally confirm a translational network composed of RNA Recognition Motif (RRM)-type RBP mRNAs that are positively controlled by HuR, another RRM-type RBP. HuR shows a preference for these RBP mRNAs bound in stem-loop motifs, confirming its role as a 'regulator of regulators'. Analysis of the transcriptome-wide HCE distribution revealed a profile of prevalently small clusters separated by unconserved intercluster RNA stretches, which predicts the formation of discrete small ribonucleoprotein complexes in the 3'-UTRs.
Figures

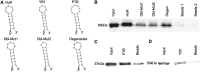
Similar articles
-
Mechanisms coordinating ELAV/Hu mRNA regulons.Curr Opin Genet Dev. 2013 Feb;23(1):35-43. doi: 10.1016/j.gde.2012.12.006. Epub 2013 Jan 9. Curr Opin Genet Dev. 2013. PMID: 23312841 Free PMC article. Review.
-
The C-terminal RNA binding motif of HuR is a multi-functional domain leading to HuR oligomerization and binding to U-rich RNA targets.RNA Biol. 2014;11(10):1250-61. doi: 10.1080/15476286.2014.996069. RNA Biol. 2014. PMID: 25584704 Free PMC article.
-
The oncogenic RNA-binding protein Musashi1 is regulated by HuR via mRNA translation and stability in glioblastoma cells.Mol Cancer Res. 2012 Jan;10(1):143-55. doi: 10.1158/1541-7786.MCR-11-0208. Mol Cancer Res. 2012. PMID: 22258704 Free PMC article.
-
Human protein-RNA interaction network is highly stable across mammals.BMC Genomics. 2019 Dec 30;20(Suppl 12):1004. doi: 10.1186/s12864-019-6330-9. BMC Genomics. 2019. PMID: 31888461 Free PMC article.
-
Functional interplay between RNA-binding protein HuR and microRNAs.Curr Protein Pept Sci. 2012 Jun;13(4):372-9. doi: 10.2174/138920312801619394. Curr Protein Pept Sci. 2012. PMID: 22708488 Free PMC article. Review.
Cited by
-
Conserved regions of the DMD 3' UTR regulate translation and mRNA abundance in cultured myotubes.Neuromuscul Disord. 2014 Aug;24(8):693-706. doi: 10.1016/j.nmd.2014.05.006. Epub 2014 May 22. Neuromuscul Disord. 2014. PMID: 24928536 Free PMC article.
-
Chimeric NP non coding regions between type A and C influenza viruses reveal their role in translation regulation.PLoS One. 2014 Sep 30;9(9):e109046. doi: 10.1371/journal.pone.0109046. eCollection 2014. PLoS One. 2014. PMID: 25268971 Free PMC article.
-
Non-coding RNAs and neuroinflammation: implications for neurological disorders.Exp Biol Med (Maywood). 2024 Feb 28;249:10120. doi: 10.3389/ebm.2024.10120. eCollection 2024. Exp Biol Med (Maywood). 2024. PMID: 38463392 Free PMC article. Review.
-
DynaMIT: the dynamic motif integration toolkit.Nucleic Acids Res. 2016 Jan 8;44(1):e2. doi: 10.1093/nar/gkv807. Epub 2015 Aug 7. Nucleic Acids Res. 2016. PMID: 26253738 Free PMC article.
-
Nutlin-Induced Apoptosis Is Specified by a Translation Program Regulated by PCBP2 and DHX30.Cell Rep. 2020 Mar 31;30(13):4355-4369.e6. doi: 10.1016/j.celrep.2020.03.011. Cell Rep. 2020. PMID: 32234473 Free PMC article.
References
-
- Mazumder B, Seshadri V, Fox PL. Translational control by the 3’-UTR: the ends specify the means. Trends Biochem. Sci. 2003;28:91–98. - PubMed
-
- Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3’UTR ends. Trends Cell Bio. 2009;19:465–474. - PubMed
-
- Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. - PubMed
-
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

