Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination
- PMID: 23377543
- PMCID: PMC3594358
- DOI: 10.1038/nsmb.2499
Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination
Abstract
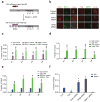
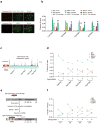
The pathogenic sequelae of BRCA1 mutation in human and mouse cells are mitigated by concomitant deletion of 53BP1, which binds histone H4 dimethylated at Lys20 (H4K20me2) to promote nonhomologous end joining, suggesting that a balance between BRCA1 and 53BP1 regulates DNA double strand-break (DSB) repair mechanism choice. Here we document that acetylation is a key determinant of this balance. TIP60 acetyltransferase deficiency reduced BRCA1 at DSB chromatin with commensurate increases in 53BP1, whereas HDAC inhibition yielded the opposite effect. TIP60-dependent H4 acetylation diminished 53BP1 binding to H4K20me2 in part through disruption of a salt bridge between H4K16 and Glu1551 in the 53BP1 Tudor domain. Moreover, TIP60 deficiency impaired homologous recombination and conferred sensitivity to PARP inhibition in a 53BP1-dependent manner. These findings demonstrate that acetylation in cis to H4K20me2 regulates relative BRCA1 and 53BP1 DSB chromatin occupancy to direct DNA repair mechanism.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





Comment in
-
DNA damage: limiting 53BP1.Nat Rev Mol Cell Biol. 2013 Mar;14(3):132. doi: 10.1038/nrm3532. Epub 2013 Feb 13. Nat Rev Mol Cell Biol. 2013. PMID: 23403718 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

