Colonization and internalization of Salmonella enterica in tomato plants
- PMID: 23377940
- PMCID: PMC3623171
- DOI: 10.1128/AEM.03704-12
Colonization and internalization of Salmonella enterica in tomato plants
Abstract
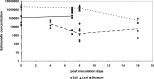
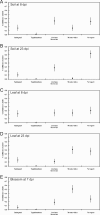
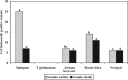
The consumption of fresh tomatoes has been linked to numerous food-borne outbreaks involving various serovars of Salmonella enterica. Recent advances in our understanding of plant-microbe interactions have shown that human enteric pathogenic bacteria, including S. enterica, are adapted to survive in the plant environment. In this study, tomato plants (Solanum lycopersicum cv. Micro-Tom) grown in sandy loam soil from Virginia's eastern shore (VES) were inoculated with S. enterica serovars to evaluate plausible internalization routes and to determine if there is any niche fitness for certain serovars. Both infested soil and contaminated blossoms can lead to low internal levels of fruit contamination with Salmonella. Salmonella serovars demonstrated a great ability to survive in environments under tomato cultivation, not only in soil but also on different parts of the tomato plant. Of the five serovars investigated, Salmonella enterica serovars Newport and Javiana were dominant in sandy loam soil, while Salmonella enterica serovars Montevideo and Newport were more prevalent on leaves and blossoms. It was also observed that Salmonella enterica serovar Typhimurium had a poor rate of survival in all the plant parts examined here, suggesting that postharvest contamination routes are more likely in S. Typhimurium contamination of tomato fruit. Conversely, S. Newport was the most prevalent serovar recovered in both the tomato rhizosphere and phyllosphere. Plants that were recently transplanted (within 3 days) had an increase in observable internalized bacteria, suggesting that plants were more susceptible to internalization right after transplant. These findings suggest that the particular Salmonella serovar and the growth stage of the plant were important factors for internalization through the root system.
Figures
References
-
- CDC 2011. Vital signs: incidence and trends of infection with pathogens transmitted commonly through food—foodborne diseases active surveillance network, 10 U.S. sites, 1996–2010. MMWR Morb. Mortal. Wkly. Rep. 60:749–755 - PubMed
-
- CDC 2011. Surveillance for foodborne disease outbreaks—United States, 2008. MMWR Morb. Mortal. Wkly. Rep. 60:1197–1202 - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

