Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities
- PMID: 23377949
- PMCID: PMC3623200
- DOI: 10.1128/AEM.03870-12
Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities
Abstract
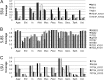
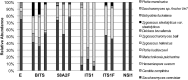
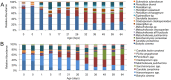
Ultra-high-throughput sequencing (HTS) of fungal communities has been restricted by short read lengths and primer amplification bias, slowing the adoption of newer sequencing technologies to fungal community profiling. To address these issues, we evaluated the performance of several common internal transcribed spacer (ITS) primers and designed a novel primer set and work flow for simultaneous quantification and species-level interrogation of fungal consortia. Primer comparison and validation were predicted in silico and by sequencing a "mock community" of mixed yeast species to explore the challenges of amplicon length and amplification bias for reconstructing defined yeast community structures. The amplicon size and distribution of this primer set are smaller than for all preexisting ITS primer sets, maximizing sequencing coverage of hypervariable ITS domains by very-short-amplicon, high-throughput sequencing platforms. This feature also enables the optional integration of quantitative PCR (qPCR) directly into the HTS preparatory work flow by substituting qPCR with these primers for standard PCR, yielding quantification of individual community members. The complete work flow described here, utilizing any of the qualified primer sets evaluated, can rapidly profile mixed fungal communities and capably reconstructed well-characterized beer and wine fermentation fungal communities.
Figures

References
-
- Bokulich NA, Bamforth CW, Mills DA. 2012. Brewhouse-resident microbiota are responsible for multi-stage fermentation of American coolship ale. PLoS One 7(4):e35507 doi:10.1371/journal.pone.0035507 - DOI - PMC - PubMed
-
- Bokulich NA, Hwang CF, Liu S, Boundy-Mills K, Mills DA. 2012. Profiling the yeast communities of wine using terminal restriction fragment length polymorphism. Am. J. Enol. Vitic. 63:185–194
-
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 - PMC - PubMed
-
- Jung MJ, Nam YD, Roh SW, Bae JW. 2012. Unexpected convergence of fungal and bacterial communities during fermentation of traditional Korean alcoholic beverages inoculated with various natural starters. Food Microbiol. 30:112–123 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

