Stable gene silencing in zebrafish with spatiotemporally targetable RNA interference
- PMID: 23378068
- PMCID: PMC3606086
- DOI: 10.1534/genetics.112.147892
Stable gene silencing in zebrafish with spatiotemporally targetable RNA interference
Abstract
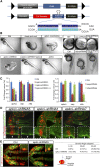
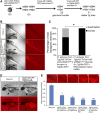
The ability to regulate gene activity in a spatiotemporally controllable manner is vital for biological discovery that will impact disease diagnosis and treatment. While conditional gene silencing is possible in other genetic model organisms, this technology is largely unavailable in zebrafish, an important vertebrate model organism for functional gene discovery. Here, using short hairpin RNAs (shRNAs) designed in the microRNA-30 backbone, which have been shown to mimic natural microRNA primary transcripts and be more effective than simple shRNAs, we report stable RNA interference-mediated gene silencing in zebrafish employing the yeast Gal4-UAS system. Using this approach, we reveal at single-cell resolution the role of atypical protein kinase Cλ (aPKCλ) in regulating neural progenitor/stem cell division. We also show effective silencing of the one-eyed-pinhead and no-tail/brachyury genes. Furthermore, we demonstrate stable integration and germ-line transmission of the UAS-miR-shRNAs for aPKCλ, the expressivity of which is controllable by the strength and expression of Gal4. This technology shall significantly advance the utility of zebrafish for understanding fundamental vertebrate biology and for the identification and evaluation of important therapeutic targets.
Figures
References
-
- Aitman T. J., Boone C., Churchill G. A., Hengartner M. O., Mackay T. F., et al. , 2011. The future of model organisms in human disease research. Nat. Rev. Genet. 12: 575–582. - PubMed
-
- Asakawa K., Kawakami K., 2010. A transgenic zebrafish for monitoring in vivo microtubule structures. Dev. Dyn. 239: 2695–2699. - PubMed
-
- Davis R. H., 2004. The age of model organisms. Nat. Rev. Genet. 5: 69–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

