PhenoDB: a new web-based tool for the collection, storage, and analysis of phenotypic features
- PMID: 23378291
- PMCID: PMC3627299
- DOI: 10.1002/humu.22283
PhenoDB: a new web-based tool for the collection, storage, and analysis of phenotypic features
Abstract
To interpret whole exome/genome sequence data for clinical and research purposes, comprehensive phenotypic information, knowledge of pedigree structure, and results of previous clinical testing are essential. With these requirements in mind and to meet the needs of the Centers for Mendelian Genomics project, we have developed PhenoDB (http://phenodb.net), a secure, Web-based portal for entry, storage, and analysis of phenotypic and other clinical information. The phenotypic features are organized hierarchically according to the major headings and subheadings of the Online Mendelian Inheritance in Man (OMIM®) clinical synopses, with further subdivisions according to structure and function. Every string allows for a free-text entry. All of the approximately 2,900 features use the preferred term from Elements of Morphology and are fully searchable and mapped to the Human Phenotype Ontology and Elements of Morphology. The PhenoDB allows for ascertainment of relevant information from a case in a family or cohort, which is then searchable by family, OMIM number, phenotypic feature, mode of inheritance, genes screened, and so on. The database can also be used to format phenotypic data for submission to dbGaP for appropriately consented individuals. PhenoDB was built using Django, an open source Web development tool, and is freely available through the Johns Hopkins McKusick-Nathans Institute of Genetic Medicine (http://phenodb.net).
© 2013 Wiley Periodicals, Inc.
Figures

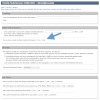
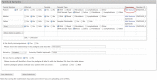
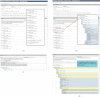
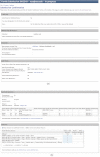
References
-
- Bamshad MJ, Shendure JA, Valle D, Hamosh A, Lupski JR, Gibbs RA, Boerwinkle E, Lifton RP, Gerstein M, Gunel M, Mane S, Nickerson DA. Centers for Mendelian Genomics. The Centers for Mendelian Genomics: a new large-scale initiative to identify the genes underlying rare Mendelian conditions. Am J Med Genet A. 2012;158A:1523–1525. - PMC - PubMed
-
- Carey JC, Allanson JE, Hennekam RC, Biesecker LG. Standard terminology for phenotypic variations: the elements of morphology project, its current progress, and future directions. Hum Mutat. 2012;33:781–786. - PubMed
-
- Guest SS, Evans CD, Winter RM. The Online London Dysmorphology Database. Genet Med. 1999;1:207–212. - PubMed
-
- Rath A, Olry A, Dhombres F, Brandt MM, Urbero B, Ayme S. Representation of rare diseases in health information systems: the Orphanet approach to serve a wide range of end users. Hum Mutat. 2012;33:803–808. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

