FOXC2 expression links epithelial-mesenchymal transition and stem cell properties in breast cancer
- PMID: 23378344
- PMCID: PMC3602160
- DOI: 10.1158/0008-5472.CAN-12-2962
FOXC2 expression links epithelial-mesenchymal transition and stem cell properties in breast cancer
Abstract
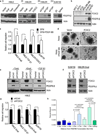
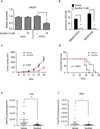
Resistance to chemotherapy and metastases are the major causes of breast cancer-related mortality. Moreover, cancer stem cells (CSC) play critical roles in cancer progression and treatment resistance. Previously, it was found that CSC-like cells can be generated by aberrant activation of epithelial-mesenchymal transition (EMT), thereby making anti-EMT strategies a novel therapeutic option for treatment of aggressive breast cancers. Here, we report that the transcription factor FOXC2 induced in response to multiple EMT signaling pathways as well as elevated in stem cell-enriched factions is a critical determinant of mesenchymal and stem cell properties, in cells induced to undergo EMT- and CSC-enriched breast cancer cell lines. More specifically, attenuation of FOXC2 expression using lentiviral short hairpin RNA led to inhibition of the mesenchymal phenotype and associated invasive and stem cell properties, which included reduced mammosphere-forming ability and tumor initiation. Whereas, overexpression of FOXC2 was sufficient to induce CSC properties and spontaneous metastasis in transformed human mammary epithelial cells. Furthermore, a FOXC2-induced gene expression signature was enriched in the claudin-low/basal B breast tumor subtype that contains EMT and CSC features. Having identified PDGFR-β to be regulated by FOXC2, we show that the U.S. Food and Drug Administration-approved PDGFR inhibitor, sunitinib, targets FOXC2-expressing tumor cells leading to reduced CSC and metastatic properties. Thus, FOXC2 or its associated gene expression program may provide an effective target for anti-EMT-based therapies for the treatment of claudin-low/basal B breast tumors or other EMT-/CSC-enriched tumors.
Figures





References
-
- Abraham BK, Fritz P, McClellan M, Hauptvogel P, Athelogou M, Brauch H. Prevalence of CD44+/CD24−/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin Cancer Res. 2005;11:1154–1159. - PubMed
-
- Liu R, Wang X, Chen GY, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–226. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

