Lamin-like analogues in plants: the characterization of NMCP1 in Allium cepa
- PMID: 23378381
- PMCID: PMC3617829
- DOI: 10.1093/jxb/ert020
Lamin-like analogues in plants: the characterization of NMCP1 in Allium cepa
Abstract
The nucleoskeleton of plants contains a peripheral lamina (also called plamina) and, even though lamins are absent in plants, their roles are still fulfilled in plant nuclei. One of the most intriguing topics in plant biology concerns the identity of lamin protein analogues in plants. Good candidates to play lamin functions in plants are the members of the NMCP (nuclear matrix constituent protein) family, which exhibit the typical tripartite structure of lamins. This paper describes a bioinformatics analysis and classification of the NMCP family based on phylogenetic relationships, sequence similarity and the distribution of conserved regions in 76 homologues. In addition, NMCP1 in the monocot Allium cepa characterized by its sequence and structure, biochemical properties, and subnuclear distribution and alterations in its expression throughout the root were identified. The results demonstrate that these proteins exhibit many similarities to lamins (structural organization, conserved regions, subnuclear distribution, and solubility) and that they may fulfil the functions of lamins in plants. These findings significantly advance understanding of the structural proteins of the plant lamina and nucleoskeleton and provide a basis for further investigation of the protein networks forming these structures.
Figures



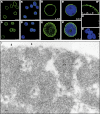
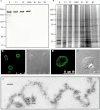

References
-
- Berkelman T. 2008. Quantitation of protein in samples prepared for 2-D electrophoresis. Methods in Molecular Biology 424, 43–49 - PubMed
-
- Blumenthal SS, Clark GB, Roux SJ. 2004. Biochemical and immunological characterization of pea nuclear intermediate filament proteins. Planta 218, 965–975 - PubMed
-
- Broers JL, Machiels BM, Kuijpers HJ, Smedts F, van den Kieboom R, Raymond Y, Ramaekers FC. 1997. A- and B-type lamins are differentially expressed in normal human tissues. Histochemistry and Cell Biology 107, 505–517 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

