Ligand binding to anion-binding exosites regulates conformational properties of thrombin
- PMID: 23378535
- PMCID: PMC3605685
- DOI: 10.1074/jbc.M112.410829
Ligand binding to anion-binding exosites regulates conformational properties of thrombin
Abstract
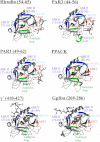
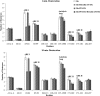
Thrombin participates in coagulation, anticoagulation, and initiation of platelet activation. To fulfill its diverse roles and maintain hemostasis, this serine protease is regulated via the extended active site region and anion-binding exosites (ABEs) I and II. For the current project, amide proton hydrogen-deuterium exchange coupled with MALDI-TOF mass spectrometry was used to characterize ligand binding to individual exosites and to investigate the presence of exosite-active site and exosite-exosite interactions. PAR3(44-56) and PAR1(49-62) were observed to bind to thrombin ABE I and then to exhibit long range effects over to ABE II. By contrast, Hirudin(54-65) focused more on ABE I and did not transmit influences over to ABE II. Although these three ligands were each directed to ABE I, they did not promote the same conformational consequences. D-Phe-Pro-Arg-chloromethyl ketone inhibition at the thrombin active site led to further local and long range consequences to thrombin-ABE I ligand complexes with the autolysis loop often most affected. When Hirudin(54-65) was bound to ABE I, it was still possible to bind GpIbα(269-286) or fibrinogen γ'(410-427) to ABE II. Each ligand exerted its predominant influences on thrombin and also allowed interexosite communication. The results obtained support the proposal that thrombin is a highly dynamic protein. The transmission of ligand-specific local and long range conformational events is proposed to help regulate this multifunctional enzyme.
Figures







References
-
- Lane D. A., Philippou H., Huntington J. A. (2005) Directing thrombin. Blood 106, 2605–2612 - PubMed
-
- Dang Q. D., Sabetta M., Di Cera E. (1997) Selective loss of fibrinogen clotting in a loop-less thrombin. J. Biol. Chem. 272, 19649–19651 - PubMed
-
- Bode W., Turk D., Karshikov A. (1992) The refined 1.9-A x-ray crystal structure of d-Phe-Pro-Arg chloromethylketone-inhibited human α-thrombin. Structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure-function relationships. Protein Sci. 1, 426–471 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

