Inhibitor of apoptosis (IAP) proteins-modulators of cell death and inflammation
- PMID: 23378585
- PMCID: PMC3552501
- DOI: 10.1101/cshperspect.a008730
Inhibitor of apoptosis (IAP) proteins-modulators of cell death and inflammation
Abstract
Misregulated innate immune signaling and cell death form the basis of much human disease pathogenesis. Inhibitor of apoptosis (IAP) protein family members are frequently overexpressed in cancer and contribute to tumor cell survival, chemo-resistance, disease progression, and poor prognosis. Although best known for their ability to regulate caspases, IAPs also influence ubiquitin (Ub)-dependent pathways that modulate innate immune signaling via activation of nuclear factor κB (NF-κB). Recent research into IAP biology has unearthed unexpected roles for this group of proteins. In addition, the advances in our understanding of the molecular mechanisms that IAPs use to regulate cell death and innate immune responses have provided new insights into disease states and suggested novel intervention strategies. Here we review the functions assigned to those IAP proteins that act at the intersection of cell death regulation and inflammatory signaling.
Figures


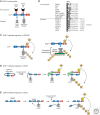



References
-
- Arama E, Agapite J, Steller H 2003. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev Cell 4: 687–697 - PubMed
-
- Bartke T, Pohl C, Pyrowolakis G, Jentsch S 2004. Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol Cell 14: 801–811 - PubMed
-
- Berger AB, Witte MD, Denault JB, Sadaghiani AM, Sexton KM, Salvesen GS, Bogyo M 2006. Identification of early intermediates of caspase activation using selective inhibitors and activity-based probes. Mol Cell 23: 509–521 - PubMed
-
- Bertrand MJ, Doiron K, Labbe K, Korneluk RG, Barker PA, Saleh M 2009. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity 30: 789–801 - PubMed
-
- Bhoj VG, Chen ZJ 2009. Ubiquitylation in innate and adaptive immunity. Nature 458: 430–437 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
