Okazaki fragment metabolism
- PMID: 23378587
- PMCID: PMC3552508
- DOI: 10.1101/cshperspect.a010173
Okazaki fragment metabolism
Abstract
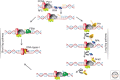
Cellular DNA replication requires efficient copying of the double-stranded chromosomal DNA. The leading strand is elongated continuously in the direction of fork opening, whereas the lagging strand is made discontinuously in the opposite direction. The lagging strand needs to be processed to form a functional DNA segment. Genetic analyses and reconstitution experiments identified proteins and multiple pathways responsible for maturation of the lagging strand. In both prokaryotes and eukaryotes the lagging-strand fragments are initiated by RNA primers, which are removed by a joining mechanism involving strand displacement of the primer into a flap, flap removal, and then ligation. Although the prokaryotic fragments are ~1200 nucleotides long, the eukaryotic fragments are much shorter, with lengths determined by nucleosome periodicity. The prokaryotic joining mechanism is simple and efficient. The eukaryotic maturation mechanism involves many enzymes, possibly three pathways, and regulation that can shift from high efficiency to high fidelity.
Figures

References
-
- Alberts BM, Barry K, Bedinger P, Formosa T, Jongeneel CV, Kreuzer KN 1982. Studies on DNA replication in the T4 bacteriophage in vitro system. Cold Spring Harb Symp Quant Biol 47: 655–668 - PubMed
-
- Ayyagari R, Gomes XV, Gordenin DA, Burgers PM 2003. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J Biol Chem 278: 1618–1625 - PubMed
-
- Bae SH, Bae KH, Kim JA, Seo YS 2001. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412: 456–461 - PubMed
-
- Balakrishnan L, Bambara RA 2011a. The changing view of Dna2. Cell Cycle 10: 2620–2621 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
