Human adipose tissue-derived mesenchymal stem cells secrete functional neprilysin-bound exosomes
- PMID: 23378928
- PMCID: PMC3561625
- DOI: 10.1038/srep01197
Human adipose tissue-derived mesenchymal stem cells secrete functional neprilysin-bound exosomes
Abstract
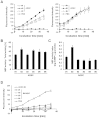
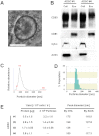
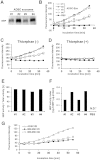
Alzheimer's disease (AD) is characterized by the accumulation of β-amyloid peptide (Aβ) in the brain because of an imbalance between Aβ production and clearance. Neprilysin (NEP) is the most important Aβ-degrading enzyme in the brain. Thus, researchers have explored virus-mediated NEP gene delivery. However, such strategies may entail unexpected risks, and thus exploration of a new possibility for NEP delivery is also required. Here, we show that human adipose tissue-derived mesenchymal stem cells (ADSCs) secrete exosomes carrying enzymatically active NEP. The NEP-specific activity level of 1 μg protein from ADSC-derived exosomes was equivalent to that of ~ 0.3 ng of recombinant human NEP. Of note, ADSC-derived exosomes were transferred into N2a cells, and were suggested to decrease both secreted and intracellular Aβ levels in the N2a cells. Importantly, these characteristics were more pronounced in ADSCs than bone marrow-derived mesenchymal stem cells, suggesting the therapeutic relevance of ADSC-derived exosomes for AD.
Figures



References
-
- Hardy J. Amyloid, the presenilins and Alzheimer's disease. Trends Neurosci. 20, 154–159 (1997). - PubMed
-
- Selkoe D. J. The cell biology of beta-amyloid precursor protein and presenilin in Alzheimer's disease. Trends Cell Biol. 8, 447–453 (1998). - PubMed
-
- Iwata N., Higuchi M. & Saido T. C. Metabolism of amyloid-beta peptide and Alzheimer's disease. Pharmacol. Ther. 108, 129–148 (2005). - PubMed
-
- Iwata N. et al. Metabolic regulation of brain Abeta by neprilysin. Science 292, 1550–1552 (2001). - PubMed
-
- Iwata N. et al. Identification of the major Abeta1-42-degrading catabolic pathway in brain parenchyma: suppression leads to biochemical and pathological deposition. Nat. Med. 6, 143–150 (2000). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

