Type II kinase inhibitors show an unexpected inhibition mode against Parkinson's disease-linked LRRK2 mutant G2019S
- PMID: 23379419
- PMCID: PMC3966205
- DOI: 10.1021/bi3012077
Type II kinase inhibitors show an unexpected inhibition mode against Parkinson's disease-linked LRRK2 mutant G2019S
Abstract
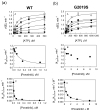
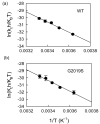
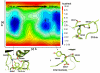
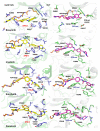
A number of well-known type II inhibitors (ATP-noncompetitive) that bind kinases in their DFG-out conformation were tested against wild-type LRRK2 and the most common Parkinson's disease-linked mutation, G2019S. We found that traditional type II inhibitors exhibit surprising variability in their inhibition mechanism between the wild type (WT) and the G2019S mutant of LRRK2. The type II kinase inhibitors were found to work in an ATP-competitive fashion against the G2019S mutant, whereas they appear to follow the expected noncompetitive mechanism against WT. Because the G2019S mutation lies in the DXG motif (DYG in LRRK2 but DFG in most other kinases) of the activation loop, we explored the structural consequence of the mutation on loop dynamics using an enhanced sampling method called metadynamics. The simulations suggest that the G2019S mutation stabilizes the DYG-in state of LRRK2 through a series of hydrogen bonds, leading to an increase in the conformational barrier between the active and inactive forms of the enzyme and a relative stabilization of the active form. The conformational bias toward the active form of LRRK2 mutants has two primary consequences. (1) The mutant enzyme becomes hyperactive, a known contributor to the Parkinsonian phenotype, as a consequence of being "locked" into the activated state, and (2) the mutation creates an unusual allosteric pocket that can bind type II inhibitors but in an ATP-competitive fashion. Our results suggest that developing type II inhibitors, which are generally considered superior to type I inhibitors because of desirable selectivity profiles, might be especially challenging for the G2019S LRRK2 mutant.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

