Multilocus sequence analysis of Treponema denticola strains of diverse origin
- PMID: 23379917
- PMCID: PMC3574001
- DOI: 10.1186/1471-2180-13-24
Multilocus sequence analysis of Treponema denticola strains of diverse origin
Abstract
Background: The oral spirochete bacterium Treponema denticola is associated with both the incidence and severity of periodontal disease. Although the biological or phenotypic properties of a significant number of T. denticola isolates have been reported in the literature, their genetic diversity or phylogeny has never been systematically investigated. Here, we describe a multilocus sequence analysis (MLSA) of 20 of the most highly studied reference strains and clinical isolates of T. denticola; which were originally isolated from subgingival plaque samples taken from subjects from China, Japan, the Netherlands, Canada and the USA.
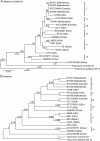
Results: The sequences of the 16S ribosomal RNA gene, and 7 conserved protein-encoding genes (flaA, recA, pyrH, ppnK, dnaN, era and radC) were successfully determined for each strain. Sequence data was analyzed using a variety of bioinformatic and phylogenetic software tools. We found no evidence of positive selection or DNA recombination within the protein-encoding genes, where levels of intraspecific sequence polymorphism varied from 18.8% (flaA) to 8.9% (dnaN). Phylogenetic analysis of the concatenated protein-encoding gene sequence data (ca. 6,513 nucleotides for each strain) using Bayesian and maximum likelihood approaches indicated that the T. denticola strains were monophyletic, and formed 6 well-defined clades. All analyzed T. denticola strains appeared to have a genetic origin distinct from that of 'Treponema vincentii' or Treponema pallidum. No specific geographical relationships could be established; but several strains isolated from different continents appear to be closely related at the genetic level.
Conclusions: Our analyses indicate that previous biological and biophysical investigations have predominantly focused on a subset of T. denticola strains with a relatively narrow range of genetic diversity. Our methodology and results establish a genetic framework for the discrimination and phylogenetic analysis of T. denticola isolates, which will greatly assist future biological and epidemiological investigations involving this putative 'periodontopathogen'.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

