Rattusin, an intestinal α-defensin-related peptide in rats with a unique cysteine spacing pattern and salt-insensitive antibacterial activities
- PMID: 23380721
- PMCID: PMC3623328
- DOI: 10.1128/AAC.02237-12
Rattusin, an intestinal α-defensin-related peptide in rats with a unique cysteine spacing pattern and salt-insensitive antibacterial activities
Abstract
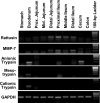
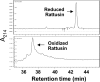
Cationic antimicrobial peptides are essential components of the innate immune system. As a major family of mammalian antimicrobial peptides, defensins are expressed mainly by mucosal epithelial cells and promyelocytes. Despite the capacity to kill a broad spectrum of bacteria through physical disruption of membranes, most defensins show substantially reduced antibacterial activities in the presence of monovalent and divalent cations, thereby limiting their therapeutic potential, particularly for the treatment of systemic infections. Genome-wide computational screening of the rat genome led to the identification of the gene for a novel α-defensin-related peptide that we termed rattusin. Rattusin shares a highly conserved signal and prosequence with mammalian α-defensins, but instead of the canonical α-defensin six-cysteine motif, rattusin consists of five cysteines with a distinctive spacing pattern. Furthermore, rattusin is preferentially expressed in Paneth cells of the distal small intestine with potent antibacterial activity against a broad range of Gram-negative and Gram-positive bacteria, including antibiotic-resistant strains. The MICs were mostly in the range of 2 to 4 μM, with no appreciable toxicity to mammalian cells at up to 100 μM. In contrast to classical α- and β-defensins, rattusin retained its activity in the presence of physiological concentrations of NaCl and Mg(2+), making it an attractive antimicrobial candidate for both topical and systemic applications.
Figures







References
-
- Finch RG. 2004. Antibiotic resistance: a view from the prescriber. Nat. Rev. Microbiol. 2:989–994 - PubMed
-
- Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389–395 - PubMed
-
- Hancock RE, Sahl HG. 2006. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24:1551–1557 - PubMed
-
- Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238–250 - PubMed
-
- Selsted ME, Ouellette AJ. 2005. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6:551–557 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

