A high-confidence interaction map identifies SIRT1 as a mediator of acetylation of USP22 and the SAGA coactivator complex
- PMID: 23382074
- PMCID: PMC3624249
- DOI: 10.1128/MCB.00971-12
A high-confidence interaction map identifies SIRT1 as a mediator of acetylation of USP22 and the SAGA coactivator complex
Abstract
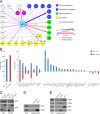
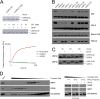
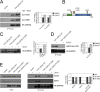
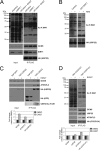
Although many functions and targets have been attributed to the histone and protein deacetylase SIRT1, a comprehensive analysis of SIRT1 binding proteins yielding a high-confidence interaction map has not been established. Using a comparative statistical analysis of binding partners, we have assembled a high-confidence SIRT1 interactome. Employing this method, we identified the deubiquitinating enzyme ubiquitin-specific protease 22 (USP22), a component of the deubiquitinating module (DUBm) of the SAGA transcriptional coactivating complex, as a SIRT1-interacting partner. We found that this interaction is highly specific, requires the ZnF-UBP domain of USP22, and is disrupted by the inactivating H363Y mutation within SIRT1. Moreover, we show that USP22 is acetylated on multiple lysine residues and that alteration of a single lysine (K129) within the ZnF-UBP domain is sufficient to alter interaction of the DUBm with the core SAGA complex. Furthermore, USP22-mediated recruitment of SIRT1 activity promotes the deacetylation of individual SAGA complex components. Our results indicate an important role of SIRT1-mediated deacetylation in regulating the formation of DUBm subcomplexes within the larger SAGA complex.
Figures





References
-
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795–800 - PubMed
-
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. U. S. A. 97:6658–6663 - PMC - PubMed
-
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. 1999. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99:735–745 - PubMed
-
- Frye RA. 1999. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 260:273–279 - PubMed
-
- Guarente L. 2000. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 14:1021–1026 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
