IDOL stimulates clathrin-independent endocytosis and multivesicular body-mediated lysosomal degradation of the low-density lipoprotein receptor
- PMID: 23382078
- PMCID: PMC3624246
- DOI: 10.1128/MCB.01716-12
IDOL stimulates clathrin-independent endocytosis and multivesicular body-mediated lysosomal degradation of the low-density lipoprotein receptor
Abstract
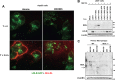
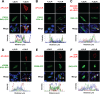
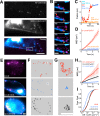
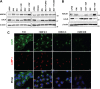
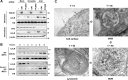
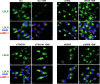
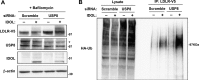
The low-density lipoprotein receptor (LDLR) is a critical determinant of plasma cholesterol levels that internalizes lipoprotein cargo via clathrin-mediated endocytosis. Here, we show that the E3 ubiquitin ligase IDOL stimulates a previously unrecognized, clathrin-independent pathway for LDLR internalization. Real-time single-particle tracking and electron microscopy reveal that IDOL is recruited to the plasma membrane by LDLR, promotes LDLR internalization in the absence of clathrin or caveolae, and facilitates LDLR degradation by shuttling it into the multivesicular body (MVB) protein-sorting pathway. The IDOL-dependent degradation pathway is distinct from that mediated by PCSK9 as only IDOL employs ESCRT (endosomal-sorting complex required for transport) complexes to recognize and traffic LDLR to lysosomes. Small interfering RNA (siRNA)-mediated knockdown of ESCRT-0 (HGS) or ESCRT-I (TSG101) components prevents IDOL-mediated LDLR degradation. We further show that USP8 acts downstream of IDOL to deubiquitinate LDLR and that USP8 is required for LDLR entry into the MVB pathway. These results provide key mechanistic insights into an evolutionarily conserved pathway for the control of lipoprotein receptor expression and cellular lipid uptake.
Figures



Similar articles
-
The LXR-IDOL axis defines a clathrin-, caveolae-, and dynamin-independent endocytic route for LDLR internalization and lysosomal degradation.J Lipid Res. 2013 Aug;54(8):2174-2184. doi: 10.1194/jlr.M037713. Epub 2013 Jun 3. J Lipid Res. 2013. PMID: 23733886 Free PMC article.
-
The Deubiquitylase USP2 Regulates the LDLR Pathway by Counteracting the E3-Ubiquitin Ligase IDOL.Circ Res. 2016 Feb 5;118(3):410-9. doi: 10.1161/CIRCRESAHA.115.307298. Epub 2015 Dec 14. Circ Res. 2016. PMID: 26666640
-
NDRG1 functions in LDL receptor trafficking by regulating endosomal recycling and degradation.J Cell Sci. 2013 Sep 1;126(Pt 17):3961-71. doi: 10.1242/jcs.128132. Epub 2013 Jun 26. J Cell Sci. 2013. PMID: 23813961
-
Post-transcriptional regulation of lipoprotein receptors by the E3-ubiquitin ligase inducible degrader of the low-density lipoprotein receptor.Curr Opin Lipidol. 2012 Jun;23(3):213-219. doi: 10.1097/MOL.0b013e3283532947. Curr Opin Lipidol. 2012. PMID: 22510808 Review.
-
Feedback regulation of cholesterol uptake by the LXR-IDOL-LDLR axis.Arterioscler Thromb Vasc Biol. 2012 Nov;32(11):2541-6. doi: 10.1161/ATVBAHA.112.250571. Epub 2012 Aug 30. Arterioscler Thromb Vasc Biol. 2012. PMID: 22936343 Free PMC article. Review.
Cited by
-
Pathways of cholesterol homeostasis in mouse retina responsive to dietary and pharmacologic treatments.J Lipid Res. 2015 Jan;56(1):81-97. doi: 10.1194/jlr.M053439. Epub 2014 Oct 7. J Lipid Res. 2015. PMID: 25293590 Free PMC article.
-
Liver X Receptor-Inducible Host E3 Ligase IDOL Targets a Human Cytomegalovirus Reactivation Determinant.J Virol. 2023 Jul 27;97(7):e0075823. doi: 10.1128/jvi.00758-23. Epub 2023 Jun 20. J Virol. 2023. PMID: 37338407 Free PMC article.
-
Engineering lentivirus envelope VSV-G for liver targeted delivery of IDOL-shRNA to ameliorate hypercholesterolemia and atherosclerosis.Mol Ther Nucleic Acids. 2024 Jan 11;35(1):102115. doi: 10.1016/j.omtn.2024.102115. eCollection 2024 Mar 12. Mol Ther Nucleic Acids. 2024. PMID: 38314097 Free PMC article.
-
PCSK9 deficiency unmasks a sex- and tissue-specific subcellular distribution of the LDL and VLDL receptors in mice.J Lipid Res. 2015 Nov;56(11):2133-42. doi: 10.1194/jlr.M061952. Epub 2015 Aug 31. J Lipid Res. 2015. PMID: 26323289 Free PMC article.
-
Dendrobium nobile Lindl. alkaloids improve lipid metabolism by increasing LDL uptake through regulation of the LXRα/IDOL/LDLR pathway and inhibition of PCSK9 expression in HepG2 cells.Exp Ther Med. 2025 Jan 9;29(3):46. doi: 10.3892/etm.2025.12796. eCollection 2025 Mar. Exp Ther Med. 2025. PMID: 39885913 Free PMC article.
References
-
- Russell DW, Schneider WJ, Yamamoto T, Luskey KL, Brown MS, Goldstein JL. 1984. Domain map of the LDL receptor: sequence homology with the epidermal growth factor precursor. Cell 37:577–585 - PubMed
-
- Brown MS, Goldstein JL. 1986. A receptor-mediated pathway for cholesterol homeostasis. Science 232:34–47 - PubMed
-
- Tolleshaug H, Hobgood KK, Brown MS, Goldstein JL. 1983. The LDL receptor locus in familial hypercholesterolemia: multiple mutations disrupt transport and processing of a membrane receptor. Cell 32:941–951 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
