A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable
- PMID: 23382174
- PMCID: PMC3616704
- DOI: 10.1093/nar/gkt041
A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable
Abstract
The human pathogen Vibrio cholerae is an aquatic bacterium associated with zooplankton and their chitinous exoskeletons. On chitinous surfaces, V. cholerae initiates a developmental programme, known as natural competence, to mediate transformation, which is a mode of horizontal gene transfer. Competence facilitates the uptake of free DNA and recombination into the bacterial genome. Recent studies have indicated that chitin surfaces are required, but not sufficient to induce competence. Two additional regulatory pathways, i.e. catabolite repression and quorum sensing (QS), are components of the regulatory network that controls natural competence in V. cholerae. In this study, we investigated the link between chitin induction and QS. We show that the major regulators of these two pathways, TfoX and HapR, are both involved in the activation of a gene encoding a transcriptional regulator of the LuxR-type family, which we named QS and TfoX-dependent regulator (QstR). We demonstrate that HapR binds the promoter of qstR in a site-specific manner, indicating a role for HapR as an activator of qstR. In addition, epistasis experiments indicate that QstR compensates for the absence of HapR. We also provide evidence that QstR is required for the proper expression of a small but essential subset of competence genes and propose a new regulatory model in which QstR links chitin-induced TfoX activity with QS.
Figures
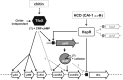
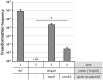

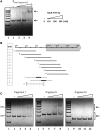
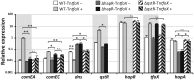
References
-
- Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. Chitin induces natural competence in Vibrio cholerae. Science. 2005;310:1824–1827. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

