Conservation and divergence in the transcriptional programs of the human and mouse immune systems
- PMID: 23382184
- PMCID: PMC3581886
- DOI: 10.1073/pnas.1222738110
Conservation and divergence in the transcriptional programs of the human and mouse immune systems
Abstract
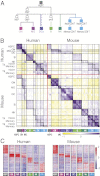
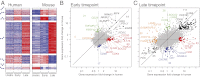
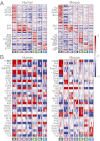
Much of the knowledge about cell differentiation and function in the immune system has come from studies in mice, but the relevance to human immunology, diseases, and therapy has been challenged, perhaps more from anecdotal than comprehensive evidence. To this end, we compare two large compendia of transcriptional profiles of human and mouse immune cell types. Global transcription profiles are conserved between corresponding cell lineages. The expression patterns of most orthologous genes are conserved, particularly for lineage-specific genes. However, several hundred genes show clearly divergent expression across the examined cell lineages, and among them, 169 genes did so even with highly stringent criteria. Finally, regulatory mechanisms--reflected by regulators' differential expression or enriched cis-elements--are conserved between the species but to a lower degree, suggesting that distinct regulation may underlie some of the conserved transcriptional responses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Payne KJ, Crooks GM. Immune-cell lineage commitment: Translation from mice to humans. Immunity. 2007;26(6):674–677. - PubMed
-
- Dermitzakis ET, Clark AG. Evolution of transcription factor binding sites in Mammalian gene regulatory regions: Conservation and turnover. Mol Biol Evol. 2002;19(7):1114–1121. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

