Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal
- PMID: 23382224
- PMCID: PMC3581968
- DOI: 10.1073/pnas.1215278110
Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal
Abstract
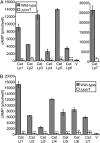
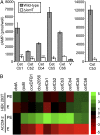
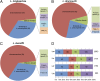
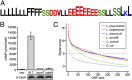
Legionella and Coxiella are intracellular pathogens that use the virulence-related Icm/Dot type-IVB secretion system to translocate effector proteins into host cells during infection. These effectors were previously shown to contain a C-terminal secretion signal required for their translocation. In this research, we implemented a hidden semi-Markov model to characterize the amino acid composition of the signal, thus providing a comprehensive computational model for the secretion signal. This model accounts for dependencies among sites and captures spatial variation in amino acid composition along the secretion signal. To validate our model, we predicted and synthetically constructed an optimal secretion signal whose sequence is different from that of any known effector. We show that this signal efficiently translocates into host cells in an Icm/Dot-dependent manner. Additionally, we predicted in silico and experimentally examined the effects of mutations in the secretion signal, which provided innovative insights into its characteristics. Some effectors were found to lack a strong secretion signal according to our model. We demonstrated that these effectors were highly dependent on the IcmS-IcmW chaperons for their translocation, unlike effectors that harbor a strong secretion signal. Furthermore, our model is innovative because it enables searching ORFs for secretion signals on a genomic scale, which led to the identification and experimental validation of 20 effectors from Legionella pneumophila, Legionella longbeachae, and Coxiella burnetii. Our combined computational and experimental methodology is general and can be applied to the identification of a wide spectrum of protein features that lack sequence conservation but have similar amino acid characteristics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Ghigo E, et al. Intracellular life of Coxiella burnetii in macrophages. Ann N Y Acad Sci. 2009;1166:55–66. - PubMed
-
- Diederen BM. Legionella spp. and Legionnaires’ disease. J Infect. 2008;56(1):1–12. - PubMed
-
- Cutler SJ, Bouzid M, Cutler RR. Q fever. J Infect. 2007;54(4):313–318. - PubMed
-
- Fields BS. The molecular ecology of legionellae. Trends Microbiol. 1996;4(7):286–290. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

