Multiple scales of selection influence the evolutionary emergence of novel pathogens
- PMID: 23382433
- PMCID: PMC3678334
- DOI: 10.1098/rstb.2012.0333
Multiple scales of selection influence the evolutionary emergence of novel pathogens
Abstract
When pathogens encounter a novel environment, such as a new host species or treatment with an antimicrobial drug, their fitness may be reduced so that adaptation is necessary to avoid extinction. Evolutionary emergence is the process by which new pathogen strains arise in response to such selective pressures. Theoretical studies over the last decade have clarified some determinants of emergence risk, but have neglected the influence of fitness on evolutionary rates and have not accounted for the multiple scales at which pathogens must compete successfully. We present a cross-scale theory for evolutionary emergence, which embeds a mechanistic model of within-host selection into a stochastic model for emergence at the population scale. We explore how fitness landscapes at within-host and between-host scales can interact to influence the probability that a pathogen lineage will emerge successfully. Results show that positive correlations between fitnesses across scales can greatly facilitate emergence, while cross-scale conflicts in selection can lead to evolutionary dead ends. The local genotype space of the initial strain of a pathogen can have disproportionate influence on emergence probability. Our cross-scale model represents a step towards integrating laboratory experiments with field surveillance data to create a rational framework to assess emergence risk.
Figures
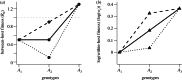
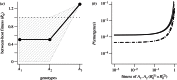

 were drawn uniformly from the ranges [1.44,1.2,1,1.01–2.24,1.02–2.25], within-host parameters N and μ as in previous figures).
were drawn uniformly from the ranges [1.44,1.2,1,1.01–2.24,1.02–2.25], within-host parameters N and μ as in previous figures).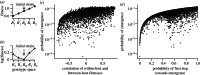
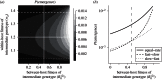
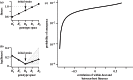
 were drawn uniformly from the ranges [1.02−2.25,1.01−2.24,1,1.01−2.24,1.02−2.25]. (c) The probability of emergence shows a positive association with the correlation between fitnesses across scales, though with considerable scatter. (d) The probability of emergence is more strongly associated with the probability that the first substitution event is towards genotype B1 (i.e. towards possible emergence). Points for plots in (c) and (d) show the results of 5000 simulated within-host landscapes. (All parameters as in figure 4.)
were drawn uniformly from the ranges [1.02−2.25,1.01−2.24,1,1.01−2.24,1.02−2.25]. (c) The probability of emergence shows a positive association with the correlation between fitnesses across scales, though with considerable scatter. (d) The probability of emergence is more strongly associated with the probability that the first substitution event is towards genotype B1 (i.e. towards possible emergence). Points for plots in (c) and (d) show the results of 5000 simulated within-host landscapes. (All parameters as in figure 4.)References
-
- Morens DM, Folkers GK, Fauci AS. 2004. The challenge of emerging and re-emerging infectious diseases. Nature 430, 242–249 (doi:10.1038/nature02759) - DOI - PMC - PubMed
-
- Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak D. 2008. Global trends in emerging infectious diseases. Nature 451, 990–993 (doi:10.1038/nature06536) - DOI - PMC - PubMed
-
- Lloyd-Smith JO, George D, Pepin KM, Pitzer VE, Pulliam JRC, Dobson AP, Hudson PJ, Grenfell BT. 2009. Epidemic dynamics at the human–animal interface. Science 326, 1362–1367 (doi:10.1126/science.1177345) - DOI - PMC - PubMed
-
- Iwasa Y, Michor F, Nowak MA. 2004. Evolutionary dynamics of invasion and escape. J. Theor. Biol. 226, 205–214 (doi:10.1016/j.jtbi.2003.08.014) - DOI - PubMed
-
- Pepin KM, Lass S, Pulliam JRC, Read AF, Lloyd-Smith JO. 2010. Identifying genetic markers of adaptation for surveillance of viral host jumps. Nat. Rev. Microbiol. 8, 802–813 (doi:10.1038/nrmicro2440) - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

