Myosin VI small insert isoform maintains exocytosis by tethering secretory granules to the cortical actin
- PMID: 23382463
- PMCID: PMC3563687
- DOI: 10.1083/jcb.201204092
Myosin VI small insert isoform maintains exocytosis by tethering secretory granules to the cortical actin
Abstract
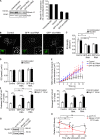
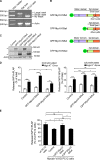
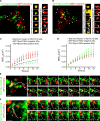
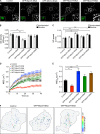
Before undergoing neuroexocytosis, secretory granules (SGs) are mobilized and tethered to the cortical actin network by an unknown mechanism. Using an SG pull-down assay and mass spectrometry, we found that myosin VI was recruited to SGs in a Ca(2+)-dependent manner. Interfering with myosin VI function in PC12 cells reduced the density of SGs near the plasma membrane without affecting their biogenesis. Myosin VI knockdown selectively impaired a late phase of exocytosis, consistent with a replenishment defect. This exocytic defect was selectively rescued by expression of the myosin VI small insert (SI) isoform, which efficiently tethered SGs to the cortical actin network. These myosin VI SI-specific effects were prevented by deletion of a c-Src kinase phosphorylation DYD motif, identified in silico. Myosin VI SI thus recruits SGs to the cortical actin network, potentially via c-Src phosphorylation, thereby maintaining an active pool of SGs near the plasma membrane.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

