Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation
- PMID: 23382536
- PMCID: PMC3638140
- DOI: 10.1101/gr.147686.112
Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation
Abstract
The detection and quantification of genetic heterogeneity in populations of cells is fundamentally important to diverse fields, ranging from microbial evolution to human cancer genetics. However, despite the cost and throughput advances associated with massively parallel sequencing, it remains challenging to reliably detect mutations that are present at a low relative abundance in a given DNA sample. Here we describe smMIP, an assay that combines single molecule tagging with multiplex targeted capture to enable practical and highly sensitive detection of low-frequency or subclonal variation. To demonstrate the potential of the method, we simultaneously resequenced 33 clinically informative cancer genes in eight cell line and 45 clinical cancer samples. Single molecule tagging facilitated extremely accurate consensus calling, with an estimated per-base error rate of 8.4 × 10(-6) in cell lines and 2.6 × 10(-5) in clinical specimens. False-positive mutations in the single molecule consensus base-calls exhibited patterns predominantly consistent with DNA damage, including 8-oxo-guanine and spontaneous deamination of cytosine. Based on mixing experiments with cell line samples, sensitivity for mutations above 1% frequency was 83% with no false positives. At clinically informative sites, we identified seven low-frequency point mutations (0.2%-4.7%), including BRAF p.V600E (melanoma, 0.2% alternate allele frequency), KRAS p.G12V (lung, 0.6%), JAK2 p.V617F (melanoma, colon, two lung, 0.3%-1.4%), and NRAS p.Q61R (colon, 4.7%). We anticipate that smMIP will be broadly adoptable as a practical and effective method for accurately detecting low-frequency mutations in both research and clinical settings.
Figures
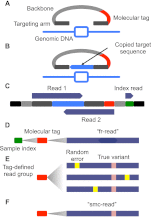
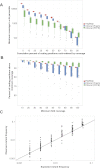

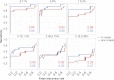
References
-
- Bielas JH, Loeb LA 2005. Quantification of random genomic mutations. Nat Methods 2: 285–290 - PubMed
-
- De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, Lamba S, Arena S, Frattini M, Piessevaux H, et al. 2010. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA 304: 1812–1820 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RC2 HL102926/HL/NHLBI NIH HHS/United States
- CA160080/CA/NCI NIH HHS/United States
- HL-102926/HL/NHLBI NIH HHS/United States
- HL-102925/HL/NHLBI NIH HHS/United States
- RC2 HL102923/HL/NHLBI NIH HHS/United States
- UC2 HL102926/HL/NHLBI NIH HHS/United States
- UC2 HL103010/HL/NHLBI NIH HHS/United States
- HL-103010/HL/NHLBI NIH HHS/United States
- HL-102924/HL/NHLBI NIH HHS/United States
- F30 AG039173/AG/NIA NIH HHS/United States
- RC2 HL102924/HL/NHLBI NIH HHS/United States
- AG039173/AG/NIA NIH HHS/United States
- UC2 HL102923/HL/NHLBI NIH HHS/United States
- UC2 HL102924/HL/NHLBI NIH HHS/United States
- RC2 HL103010/HL/NHLBI NIH HHS/United States
- T32 GM007266/GM/NIGMS NIH HHS/United States
- HL-102923/HL/NHLBI NIH HHS/United States
- R21 CA160080/CA/NCI NIH HHS/United States
- RC2 HL102925/HL/NHLBI NIH HHS/United States
- UC2 HL102925/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
