'Overgrowth' mutants in barley and wheat: new alleles and phenotypes of the 'Green Revolution' DELLA gene
- PMID: 23382550
- PMCID: PMC3617830
- DOI: 10.1093/jxb/ert022
'Overgrowth' mutants in barley and wheat: new alleles and phenotypes of the 'Green Revolution' DELLA gene
Abstract
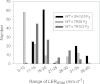
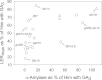
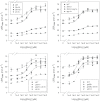
A suppressor screen using dwarf mutants of barley (Hordeum vulgare L.) led to the isolation of 'overgrowth' derivatives, which retained the original dwarfing gene but grew at a faster rate because of a new mutation. The new mutations were in the Slender1 (Sln1) gene (11/13 cases), which encodes the DELLA protein central to gibberellin (GA) signalling, showed 100% genetic linkage to Sln1 (1/13), or were in the Spindly1 (Spy1) gene (1/13), which encodes another protein involved in GA signalling. The overgrowth mutants were characterized by increased GA signalling, although the extent still depended on the background GA biosynthesis capacity, GA receptor function, and DELLA activity. A comparison between two GA responses, α-amylase production and leaf growth rate, revealed degrees of specificity for both the overgrowth allele and the GA response under consideration. Many overgrowth mutants were also isolated in a dwarf line of bread wheat (Triticum aestivum L.) and 19 new alleles were identified in the Rht-B1 gene, one of the 'Green Revolution' semi-dwarfing genes and the orthologue of Sln1. The sites of amino acid substitutions in the DELLA proteins of both species provide insight into DELLA function, and included examples where identical but independent substitutions were observed. In both species, the starting lines were too dwarfed to be directly useful in breeding programmes, but new overgrowth derivatives with semidwarf heights have now been characterized. The variation they exhibit in GA-influenced traits identifies novel alleles with perfect markers that are of potential use in breeding.
Figures



References
-
- Asano K, Hirano K, Ueguchi-Tanaka M, Angeles-Shim RB, Komura T, Satoh H, Kitano H, Matsuoka M, Ashikari M. 2009. Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice. Molecular Genetics and Genomics 281, 223–231 - PubMed
-
- Carol P, Peng J, Harberd NP. 1995. Isolation and preliminary characterization of gas1-1, a mutation causing partial suppression of the phenotype conferred by the gibberellin-insensitive (gai) mutation in Arabidopsis thaliana (L.) Heyhn. Planta 197, 414–417 - PubMed
-
- Chandler PM, Harding CA, Ashton AR, Mulcair MD, Dixon NE, Mander LN. 2008. Characterization of gibberellin receptor mutants of barley (Hordeum vulgare L.). Molecular Plant 1, 285–294 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

