Dissection of Ire1 functions reveals stress response mechanisms uniquely evolved in Candida glabrata
- PMID: 23382685
- PMCID: PMC3561209
- DOI: 10.1371/journal.ppat.1003160
Dissection of Ire1 functions reveals stress response mechanisms uniquely evolved in Candida glabrata
Abstract
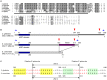
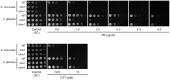
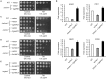
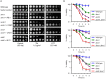
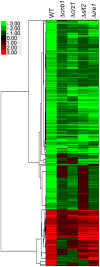
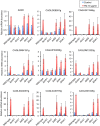
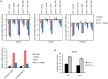
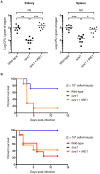
Proper protein folding in the endoplasmic reticulum (ER) is vital in all eukaryotes. When misfolded proteins accumulate in the ER lumen, the transmembrane kinase/endoribonuclease Ire1 initiates splicing of HAC1 mRNA to generate the bZIP transcription factor Hac1, which subsequently activates its target genes to increase the protein-folding capacity of the ER. This cellular machinery, called the unfolded protein response (UPR), is believed to be an evolutionarily conserved mechanism in eukaryotes. In this study, we comprehensively characterized mutant phenotypes of IRE1 and other related genes in the human fungal pathogen Candida glabrata. Unexpectedly, Ire1 was required for the ER stress response independently of Hac1 in this fungus. C. glabrata Ire1 did not cleave mRNAs encoding Hac1 and other bZIP transcription factors identified in the C. glabrata genome. Microarray analysis revealed that the transcriptional response to ER stress is not mediated by Ire1, but instead is dependent largely on calcineurin signaling and partially on the Slt2 MAPK pathway. The loss of Ire1 alone did not confer increased antifungal susceptibility in C. glabrata contrary to UPR-defective mutants in other fungi. Taken together, our results suggest that the canonical Ire1-Hac1 UPR is not conserved in C. glabrata. It is known in metazoans that active Ire1 nonspecifically cleaves and degrades a subset of ER-localized mRNAs to reduce the ER load. Intriguingly, this cellular response could occur in an Ire1 nuclease-dependent fashion in C. glabrata. We also uncovered the attenuated virulence of the C. glabrata Δire1 mutant in a mouse model of disseminated candidiasis. This study has unveiled the unique evolution of ER stress response mechanisms in C. glabrata.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
The protein kinase Ire1 has a Hac1-independent essential role in iron uptake and virulence of Candida albicans.PLoS Pathog. 2022 Feb 2;18(2):e1010283. doi: 10.1371/journal.ppat.1010283. eCollection 2022 Feb. PLoS Pathog. 2022. PMID: 35108336 Free PMC article.
-
ER stress response mechanisms in the pathogenic yeast Candida glabrata and their roles in virulence.Virulence. 2014 Feb 15;5(2):365-70. doi: 10.4161/viru.27373. Epub 2013 Dec 11. Virulence. 2014. PMID: 24335436 Free PMC article. Review.
-
Unique evolution of the UPR pathway with a novel bZIP transcription factor, Hxl1, for controlling pathogenicity of Cryptococcus neoformans.PLoS Pathog. 2011 Aug;7(8):e1002177. doi: 10.1371/journal.ppat.1002177. Epub 2011 Aug 11. PLoS Pathog. 2011. PMID: 21852949 Free PMC article.
-
Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response.Proc Natl Acad Sci U S A. 2013 Apr 2;110(14):5713-8. doi: 10.1073/pnas.1219047110. Epub 2013 Mar 18. Proc Natl Acad Sci U S A. 2013. PMID: 23509268 Free PMC article.
-
The Unfolded Protein Response Pathway in the Yeast Kluyveromyces lactis. A Comparative View among Yeast Species.Cells. 2018 Aug 14;7(8):106. doi: 10.3390/cells7080106. Cells. 2018. PMID: 30110882 Free PMC article. Review.
Cited by
-
Engineering of the unfolded protein response pathway in Pichia pastoris: enhancing production of secreted recombinant proteins.Appl Microbiol Biotechnol. 2021 Jun;105(11):4397-4414. doi: 10.1007/s00253-021-11336-5. Epub 2021 May 26. Appl Microbiol Biotechnol. 2021. PMID: 34037840 Free PMC article. Review.
-
The fungal UPR: a regulatory hub for virulence traits in the mold pathogen Aspergillus fumigatus.Virulence. 2014 Feb 15;5(2):334-40. doi: 10.4161/viru.26571. Epub 2013 Oct 18. Virulence. 2014. PMID: 24189125 Free PMC article. Review.
-
The protein kinase Ire1 has a Hac1-independent essential role in iron uptake and virulence of Candida albicans.PLoS Pathog. 2022 Feb 2;18(2):e1010283. doi: 10.1371/journal.ppat.1010283. eCollection 2022 Feb. PLoS Pathog. 2022. PMID: 35108336 Free PMC article.
-
Calcineurin as a Multifunctional Regulator: Unraveling Novel Functions in Fungal Stress Responses, Hyphal Growth, Drug Resistance, and Pathogenesis.Fungal Biol Rev. 2014 Oct;28(2-3):56-69. doi: 10.1016/j.fbr.2014.02.004. Fungal Biol Rev. 2014. PMID: 25383089 Free PMC article.
-
IreA Controls Endoplasmic Reticulum Stress-Induced Autophagy and Survival through Homeostasis Recovery.Mol Cell Biol. 2018 Jun 14;38(13):e00054-18. doi: 10.1128/MCB.00054-18. Print 2018 Jul 1. Mol Cell Biol. 2018. PMID: 29632077 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

